Abstract
The Forkhead transcription factors FoxO1, FoxO3a, and FoxO4 play a prominent role in regulating cell survival and cell cycle. Whereas FOXO1 was shown to mediate insulin sensitivity and adipocyte differentiation, the role of the transcription factor FoxO4 in metabolism remains ill defined. To uncover the effects of FoxO4, we generated a cellular model of stable FoxO4 overexpression and subjected it to microarray-based gene expression profiling. While pathway analysis revealed a disruption of cholesterol biosynthesis gene expression, biochemical studies revealed an inhibition of cholesterol biosynthesis, which was coupled with decreased mRNA levels of lanosterol 14α demethylase (CYP51). FoxO4-mediated repression of CYP51 led to the accumulation of 24,25 dihydrolanosterol (DHL), which independently and unlike lanosterol inhibited cholesterol biosynthesis. Furthermore, FoxO4-overexpressing cells accumulated lipid droplets and triacylglycerols and had an increase in basal glucose uptake. Recapitulation of these effects was obtained following treatment with CYP51 inhibitors, which also induce DHL buildup. Moreover, DHL but not lanosterol strongly stimulated liver X receptor α (LXRα) activity, suggesting that DHL and LXRα mediate the downstream effects initiated by FoxO4. Together, these studies suggest that FoxO4 acts on CYP51 to regulate the late steps of cholesterol biosynthesis.
Keywords: CYP51, dihydrolanosterol, lanosterol, liver X receptor α, miconazole, ketoconazole
The initial clue between forkhead proteins (1) and metabolism originated from the nematode Caenorhabditis elegans when it was found that inactivation of the insulin receptor homolog daf-2 shifted the metabolism of the worm to fat storage, resulting in a prolonged life span (2). The O family of the forkhead transcription factors FoxO1, FoxO3a, and FoxO4 have been involved in the regulation of multiple biological pathways (3), such as cell survival (4), cell cycle (5), insulin sensitivity (6), and adipocyte differentiation (7). FoxO transcription factors are expressed at different levels in multiple tissues, with FoxO4 showing predominant expression in skeletal muscle, heart, and adipose tissue (1, 8). Mouse knockout studies of FoxO1, FoxO3a, and FoxO4 revealed that homozygosity for FoxO1 led to embryonic lethality, whereas FoxO3a and FoxO4 were viable (9). These studies demonstrate that FoxO3a and FoxO4 cannot compensate for the loss of FoxO1; however, FoxO1 or other factors could compensate for certain aspects of FoxO3a and FoxO4 loss of function. FoxO3a homozygous null females showed an age-dependent infertility associated with abnormal follicular development, and transgenic mice overexpressing a constitutively active form of FoxO3a in oocytes had delayed oocyte growth and follicular development (9, 10). Although FoxO4 homozygous null mice had no overt phenotype (9), recent studies have shown that they were more susceptible to injury-induced colitis and increased intestinal epithelial permeability (11).
An overexpression model of FoxO4 while bypassing issues of gene compensation arising from knockout studies might uncover new pathways associated with FoxO4 and could reveal differential modes of FoxO4 regulation. In this vein, we recently determined that transgenic mice overexpressing a constitutively active form of FoxO4 in adipose tissue have rapid glucose clearance (12). In this report, we demonstrate that in an overexpression cellular model, FoxO4 inhibits the late steps of cholesterol biosynthesis, leading to the accumulation of 24,25 dihydrolanosterol (DHL), which impacts triacylglycerol (TAG) accumulation and basal glucose uptake, likely through the stimulation of the liver X receptor α (LXRα).
MATERIALS AND METHODS
Generation and characterization of 3T3L1 stable cell lines
Plasmid pBabe vectors harboring the puromycin gene alone (Puro) or a single hemaglutinin tagged to human FoxO4 (HA-FoxO4) coding sequences were transfected into the mouse CREBAG2 packaging cell line using Lipofectamine 2000 (Invitrogen). Seventy-two hours later, retroviruses shed in the culture medium were harvested from CREBAG2 supernatants every 8 h and stored in culture medium at −80°C or used immediately for transduction. The 3T3L1 fibroblasts grown in 10 cm plates were transduced overnight with either Puro or HA-FoxO4 retroviruses in 2 ml of DMEM supplemented with 0.5 g/l glucose, 10% newborn calf serum (DMEM/gluc/serum), and 6 μg/ml polybrene. Transduced cells were cultured in DMEM/gluc/serum for 72 h before selection in the same medium containing 2 μg/ml puromycin. Puromycin-resistant cells were allowed to form ring colonies before being transferred to individual 6-well plates for further growth in puromycin selective medium.
Western blots, real-time PCR, and staining of lipid droplets
Puromycin-resistant clones were analyzed for HA-FoxO4 protein expression by Western blot analysis and for HA-FoxO4 and endogenous FoxO4 mRNA expression by real-time quantitative PCR (qPCR). Total protein lysates were prepared and used for Western blots as previously described with 20 μg protein loaded into each lane (12). Antibodies to HA, α-tubulin, and FoxO4 (N19) were obtained from Santa Cruz Biochemicals (Santa Cruz, CA) and Akt and phospho Akt 473 from Cell Signaling Technologies (Beverly, MA). Nuclear and cytoplasmic extracts were prepared from Puro and HA-FoxO4 cells with a kit from Active Motif (Carlsbad, CA). Nuclear and cytoplasmic protein bands were quantitated with NIH ImageJ software (version 10.2) to determine the nuclear to cytoplasmic ratios.
PCR primers for HA-FoxO4 were 5′-GCTTACCCATACGATGTTCCA-3′ and 5′-TGTGTACCTTTTCCCCCAGA-3′, for endogenous FoxO4 expression 5′-TCATCAAGGTTCACAACGAGGC-3′ and 5′-AGGACAGACGGCTTCTTCTTGG-3′, and for the cyclophilin internal control mRNA 5′-ATAATGGCACTGGCGGCAGG-3′ and 5′-CCATCCAGCCATTCAGTCTTGG-3′.
To visualize lipid droplets, live adherent cells were stained for 10 min in PBS with 0.5 μg/ml Nile Red (from Sigma, St. Louis, MO) prepared from a 0.5 mg/ml stock solution in acetone. Cells were then washed with PBS and the lipid droplets visualized and imaged on a fluorescent microscope. All experiments were performed on confluent cells grown in 10 cm dishes or 6-well plates.
Luciferase assays
The pGL3 plasmid containing three copies of the forkhead-responsive element (FRE) linked to the firefly luciferase reporter gene was obtained from the laboratory of Dr. Michael Greenberg via Addgene (Cambridge, MA; plasmid 1789). Two hundred nanograms of this plasmid was cotransfected with the same amount of either pBabe Puro or pBabe HA-FoxO4 into HEK293 cells with the lipid-based reagent Superfect (Qiagen, Germantown, MD) according to the manufacturer's protocol. Transfections also included 20 ng of the renilla luciferase plasmid as internal control for the assay. Cells were harvested 24 h after transfection and luciferase assays performed with a kit from Invitrogen (Carlsbad, CA). Fluorescence was read on a Turner Biosystems luminometer. All transfections were done in triplicate wells, and the average fluorescence of firefly-to-renilla luciferase ratios calculated. HA-FoxO4 activity ratios were normalized to those of Puro controls.
Microarray and qPCR studies
Two groups of Puro and HA-FoxO4 cells were grown to confluence in 10 cm dishes in standard medium (DMEM/glucose, 10% newborn calf serum). One group was serum starved for 8 h (DMEM only) and the other maintained in DMEM/10% serum. RNA was then extracted with the Trizol reagent from five individual plates in each group. RNA pools were prepared by mixing equal RNA amounts from each group. Total pooled RNA from the four groups (Puro fed, HA-FoxO4 fed, Puro serum starved, and HA-FoxO4 serum starved) was used as a template for cDNA synthesis, dye labeling, and hybridization to Affymetrix mouse microarray 2.0 chips. Pathway analysis of the data was performed with the GeneSifter software, which generated the Z score representing the number of standard deviations from the mean for each pathway and normalized gene expression levels.
qPCR was performed on individual RNAs extracted from Puro and HA-FoxO4 cells grown in 10% serum or in its absence for 8 h. PCR primers aimed at the amplification of mRNA from six representative cholesterol biosynthetic genes (HMG-CoA reductase, HMG-CoA synthase, isopentenyl diphosphate isomerase, sterol regulatory protein 2, 24-dehydrocholesterol reductase, and lanosterol 14α demethylase) are listed in Table 1. qPCR was performed with SYBR Green PCR mixes on MJ Research/Bio-Rad thermal cyclers. The Ct values obtained from the linear portion of the amplification curves of each gene, the cyclophilin internal control, and their respective amplification efficiency slopes were exported to the QGENE Excel template for mRNA relative quantitation and statistical analysis.
TABLE 1.
DNA sequence of PCR primers used for real-time amplification of selected genes in cholesterol biosynthesis
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| HMG-CoA synthase | 5′-TGGAGACCACAGTTCTCTGTCCTT-3′ | 5′-GCAACGATTCCCACATCTTTTGGC-3′ |
| HMG-CoA reductase | 5′-GCGAGTGCATTAGCAAAGTTTGCC-3′ | 5′-GCATTCCACAAGAGCGTCAAGAGT-3′ |
| Isopentenyl diphosphate isomerase | 5′-CGCTTGAAAGCCGAGTTGGGAATA-3′ | 5′-TGTTCACCCCAGATACCATCAGA-3′ |
| SREBP2 | 5′-AGCTGTGCGCTCTCGTTTTACT-3′ | 5′-AGGCATGCATGGCTCTACAGGTAT-3′ |
| Δ24 sterol reductase | 5′-GCTGCGAGTCGGAAAGTACAAGAA-3′ | 5′-TGTCACCTGACCCATAGACACCAA-3′ |
| Lanosterol demethylase (CYP51) | 5′-ACGCAGACCTGGATGGAGGTTTTA-3′ | 5′-TCTTTTGACAGCCTGCGCTTCT-3′ |
Lipids biosynthesis, analysis, and mass spectroscopy
De novo lipid biosynthesis was assessed by 14C-acetate (GE Healthcare/Amersham; 57 mCi/mmol, 0.2 μCi/μl) incorporation assays in cells grown in 6-well plates. Briefly, cells were serum starved for 2 h in 1 ml DMEM/glucose followed by the addition of 1 μCi of 14C-acetate and incubated for 4 h at 37°C, 5% CO2. In some experiments, cells were pretreated for 30 min with either 50 μM LY 294002 (PI3-kinase inhibitor), 50 μM PD9805 [a mitogen-activated protein (MAP) kinase inhibitor], 0.1 μM rapamycin (mTOR inhibitor), 10 μM cerulenin (a fatty acid biosynthesis inhibitor), 20 μg/ml lovastatin (a cholesterol biosynthesis inhibitor), 5 μM H89 (protein kinase A inhibitor), 20 μM compound C (AMP kinase inhibitor), or 10 μM cytochalasin B (glucose transport inhibitor). After 4 h of incubation, cells were washed twice with PBS and recovered in 0.2 ml of PBS. Lipids were extracted (13) and fractionated by TLC on channeled Whatman 250 μm silica plates in a mobile phase consisting of hexane:ether:acetic acid (80:20:1). Visualization of lipids was with iodine vapor followed, if appropriate, by exposure to X-ray films for 3–10 days or overnight exposure to a phosphor imager screen. Densitometric scanning and analysis of signal intensity was performed with NIH ImageJ software.
For preparative analysis, cells grown in 10 cm dishes were recovered each as a 0.5 ml suspension in PBS followed by lipids extraction and fractionation as above except that the TLC plates were uniformly coated with silica. The unknown lipid band was recovered by scraping, extracted with 2:1 methanol:chloroform, and dried in Teflon capped vials for mass spectroscopy analysis. All lipid markers were from Sigma (St. Louis, MO), except dihydrolanosterol, which was from Steraloids (Newport, RI).
Gas chromatography and mass spectroscopy
Identification of unknown lipids was performed by GC-selected ion monitoring-MS at the Kansas Lipidomics Research Center (Kansas State University, Manhattan, KS). All samples were derivatized. GC analysis of the sterol samples was performed on an Agilent 6890N coupled to an Agilent model 5975N quadrupole mass selective detector. Separation was achieved on an HP-5MS fused silica capillary column (5% phenyl methyl siloxane; column length, 30 m; internal diameter, 0.25 mm; film thickness, 0.25 μm). The GC operating conditions were as follows: initial temperature of 50°C, holding time of 5 min, increasing to 310°C at a rate of 20°C/min, with a final isothermal hold at 310°C for 15 min. Helium was used as carrier gas. The samples (1 μl) were injected in a split (5:1) mode with an Agilent 7683 autosampler. The injector temperature was set at 275°C, and the mass spectrometry transfer line was at 280°C. The mass spectrometer was operated in the electron impact mode at 70 eV ionization energy and scanned from 50 to 650 Da. The temperatures of the MS quadrupole and source were set at 150°C and 250°C, respectively. Data were acquired and processed with Agilent Chemstation software. Individual compounds were identified by comparison of mass spectra with the NIST Mass Spectral library. Analysis of each sample was performed in triplicate (three injections of each derivatized sample).
Lipids and fatty acids analysis of Puro and HA-FoxO4 cells
Eight million cells from each Puro and HA-FoxO4 were grown under serum-fed conditions, trypsinized, recovered as cell pellets, and frozen in liquid nitrogen. The resulting lipid extracts were analyzed by HPLC and GC flame ionization at Lipomics Technologies (West Sacramento, CA). The analysis yielded the major lipid classes and the fatty acids constituents of TAGs and cholesteryl esters.
3H-deoxyglucose uptake assays
3H-deoxy-d-glucose (8 Ci/mmol, 1 μCi/μl) was obtained from Perkin-Elmer (Boston, MA). Confluent cells grown in 6-well plates were incubated for 2 h in KRHB buffer (0.12 M NaCl, 4 mM KH2PO4, 1 mM MgSO4, 0.75 mM CaCl2, 10 mM NaHCO3, and 30 mM HEPES, pH 7.4) with or without 50 or 100 nM insulin, followed by the addition of 50 μM 2-deoxyglucose (2DG; from Sigma) and 0.5 μCi 3H-2DG. Cells were incubated for further 30 min at 37°C, 5% CO2, washed twice with ice-cold PBS, then lysed with 0.5 ml 50 mM NaOH/1% Triton X-100. A 200 μl aliquot from each well was used to determine the incorporated radioactivity by liquid scintillation counting. The incorporated radioactivity was normalized to the protein content of cells in each well. Protein assays were performed by the microBCA assay (Pierce, Rockford, IL). In some experiments, pretreatment of the cells with insulin, 50 μM LY 294002, 50 μM PD9805, 1 μM rapamycin, or 10 μM cytochalasin B (all obtained from Calbiochem/EMD Biosciences) was carried out for 30 min prior to addition of 2DG and 3H-2DG.
Treatment of cells with miconazole, ketoconazole, lanosterol, or DHL
Cells in 6-well plates were treated with vehicle (0.1% DMSO) and different concentrations of miconazole or ketoconazole (Sigma) for 1 week to induce sterol accumulation. Lipids extracts and TLC analysis were performed as described above. Determination of TAG levels in lipid extracts was performed with the GPO Trinder assay (Pointe Scientific, Canton, MI) on dried aliquots from the lipid extracts or dilutions of a glycerol standard (Sigma) for generation of a standard curve. 3H-2DG assays were performed as described above. Protein assays were performed as described above on protein precipitates recovered from lipid extracts and dissolved in 10 mM NaOH and 0.5% Triton X-100.
The purities of lanosterol (Sigma; L5768) and DHL (Steraloids, Newport, RI) were established by GC-MS at the Kansas Lipidomics Research Center and consisted each of 99% purity with no other traces of sterols. Lanosterol and DHL were dissolved each in DMSO as a 7.7 mM stock solution. An aliquot of each sterol was added to the culture media of 3T3L1 fibroblasts or Hepa1-6 hepatocytes (hepatoma cell line obtained from ATCC) to a final concentration of 10 μM or 40 μM. Pretreatment of cells with each sterol was for 1 h in DMEM/glucose followed by the addition of 1 μCi of 14C acetate and incubation for additional 4 h at 37°C/5%CO2. Following lipids extraction, TLC fractionation, iodine vapor staining, and autoradiography, individual lipid bands were scraped into scintillation vials and counted. The determined radioactivity was normalized to the protein content recovered from each lipid extract.
Coactivator recruitment assay
Fluorescence resonance energy transfer (FRET) assay was performed in 384-well plates in 20 μl of volume. A mix of 5 nM His-LXRαLBD or His-LXRβLBD (Roche), 5 nM biotinylated SRC-1 peptide (Biotin-CPSSHSSLTERHKILHRLLQEGSPSC-OH), Europium-labeled anti-His antibody (Perkin-Elmer), and allophycocyanin-labeled streptavidin (Prozyme) was prepared. The 24(S),25 epoxycholesterol (EPCH), DHL, and lanosterol were tested in 12 point dose-response curves starting at 100 μM. Plates were incubated for 3 h at room temperature and FRET read on an AnalystGT (Molecular Devices). Compound dose responses were run in duplicate. Dose response curves were generated by nonlinear regression analysis using GraphPad Prism.
LXR transcriptional assays
Transient transfection assays were carried out as previously described (14). Briefly, an expression construct for a chimeric GAL and human LXRα ligand binding domain (LBD) was generated by fusing the human LXRα to the DNA binding domain of the yeast transcription factor GAL4 and inserting it into pcDNA3 (Invitrogen). The 10T 1/2 cells were cultured in DMEM with 10% FBS. For transcriptional assays, cells were plated in subconfluent conditions and transfected with the GAL-LXRα LBD expression construct and an upstream activating sequence luciferase reporter using FuGENE 6 (Roche). Twenty-four hours later, cells were switched to serum-free DMEM and treated for 6 h with the synthetic LXR ligand GW3965 (1 µM), 24(S),25 epoxycholesterol (9 µM), lanosterol (9 µM), or DHL (9 µM). Luciferase activity was read using BriteLite reagent (Perkin-Elmer).
RESULTS
Generation of HA-FoxO4-overexpressing cell lines
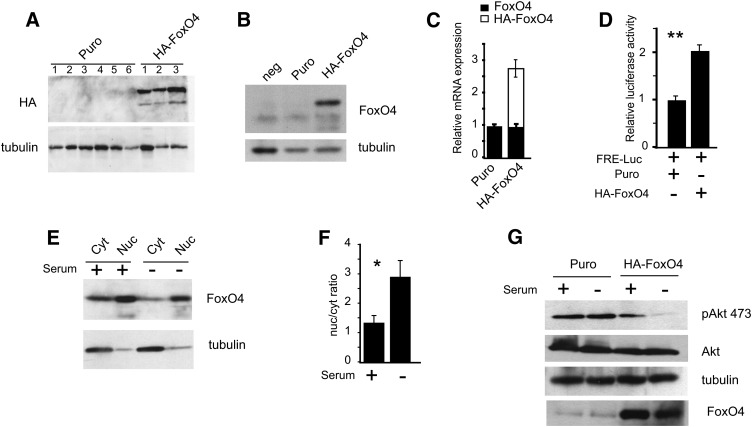
Puromycin-resistant clone isolates overexpressing HA-FoxO4 were confirmed for protein expression by Western blot analysis from corresponding protein lysates with HA- or FoxO4-specific antibodies (Fig. 1A, B). Cells from pBabe Puro-transduced retroviruses were consistently negative for HA-FoxO4 expression, thus providing a negative control cell line for subsequent studies. Puro and HA-FoxO4 cells were used as mixed pools in all experiments in order to reduce unique effects associated with individual clones. qPCR revealed that HA-FoxO4 mRNA overexpression consisted of 2.8-fold higher than endogenous FoxO4 mRNA levels (Fig. 1C), demonstrating moderately elevated levels of HA-FoxO4 expression relative to endogenous levels.
Fig. 1.
Derivation of stable 3T3L1 fibroblasts expressing a hemaglutinin-tagged (HA) form of FoxO4. A: Western blot analysis of protein extracts from stable cell lines transduced with pBabe Puro or pBabe HA-FoxO4 retroviruses. HA-FoxO4 was detected with anti-HA, while anti-tubulin was used to reveal protein loading. The Western blot shows six negative and three positive cell lines. B: Western blot with anti-FoxO4 from protein extracts of two clones, a Puro negative control, and HA-FoxO4. The protein band below HA-FoxO4 is likely to represent endogenous FoxO4 and appears in all three lanes. C: Expression levels by real-time qPCR of endogenous FoxO4 and HA-FoxO4 mRNA in Puro and HA-FoxO4 cell lines (n = 6 in each group). D: Cotransfection of FRE-luciferase with either Puro or HA-FoxO4. Relative luciferase activity is shown for three independent experiments. Asterisks indicate P < 0.01. E: Western blot of nuclear and cytoplasmic protein extracts from HA-FoxO4 cells either serum fed (+) or starved (−). The blots were conjugated with either anti-FoxO4 or anti-tubulin. A small amount of cytoplasmic tubulin is carried over to the nuclear fraction. F: Nuclear-to-cytoplasmic (nuc/cyt) ratio of HA-FoxO4 in serum fed (+) and starved (−) conditions as determined by densitometric scan of the Western blot shown in E. Asterisk indicates P < 0.05. G: Western blot analysis of protein lysates from Puro and HA-FoxO4 cells either serum fed (+) or starved (−) and conjugated to anti-phosphoserine 473 Akt, anti-Akt, anti-tubulin (for protein loading control), or anti-FoxO4.
The ability of HA-FoxO4 to bind to FREs and stimulate gene expression was demonstrated by FRE-luciferase transfection assays. We found that HA-FoxO4 induced by 2-fold luciferase activity (Fig. 1D). Furthermore, HA-FoxO4 was predominantly localized in the nucleus with a 2.2-fold (P < 0.03) more abundance in the nucleus than in the cytoplasm under serum withdrawal conditions (Fig. 1E, F), which are also associated with reduced phosphorylation of protein kinase B/akt in HA-FoxO4 cells (Fig. 1G).
HA-FoxO4 disrupts cholesterol biosynthesis gene expression
Pathway analysis of gene expression profiling of Puro and HA-FoxO4 (Table 2) cells revealed that sterol and steroid biosynthesis/metabolism were overrepresented with Z scores ranging from 11.9–8.7. Mitosis and cell cycle pathways, which are regulated by FoxO4 (5), constituted the next most significant pathways with Z scores of 8.3–8.4. Lipid, and more specifically fatty acid, biosynthesis pathways were also elevated in HA-FoxO4 cells, albeit at lower significance than cholesterol biosynthesis. Consistent with the overrepresentation of cholesterol and fatty acids metabolism, the microarray data also showed in HA-FoxO4 cells an increase in the relative expression levels of genes in these two pathways.
TABLE 2.
Pathways and lipid metabolism genes impacted by the overexpression of HA-FoxO4 in serum starved Puro versus HA-FoxO4 cells
| Ontology Group | Z Score |
|---|---|
| Sterol biosynthesis | 11.9 |
| Sterol metabolism | 10.6 |
| Cholesterol biosynthesis | 8.8 |
| Steroid biosynthesis | 8.7 |
| Cholesterol metabolism | 8.5 |
| Mitotic cell cycle | 8.4 |
| Mitosis | 8.4 |
| Positive regulation of progression through cell cycle | 8.3 |
| M phase of mitotic cell cycle | 8.3 |
| Lipid biosynthesis | 5.5 |
| Fatty acid elongation | 4.0 |
| Fatty acid desaturation | 3.0 |
| Fatty acid biosynthesis | 2.8 |
| Cholesterol Biosynthesis | Fold increase in HA-FoxO4 vs. Puro |
| Hydroxymethylglutaryl CoA synthase (HMGCS1) | 10.9 |
| Hydroxymethylglutaryl CoA reductase (HMGCR) | 9.9 |
| Isopentenyl diphosphate δ isomerase (IPDI) | 39.2 |
| Farnesyl diphospho transferase (FDFS1) | 6.3 |
| NADP-dependent steroid dehydrogenase (NADPSD) | 5.8 |
| Δ-24 sterol reductase (DHCR24) | 3.4 |
| Lanosterol 14α demethylase (CYP51) | 1.3 |
| SREBP2 | 3.6 |
| Fatty Acid Biosynthesis, Elongation, and Desaturation | |
| Acetyl-CoA Carboxylase (ACC1) | 1.6 |
| Fatty Acid Synthase (FASN) | 3.1 |
| Long-chain fatty-acyl elongase (ELOVL6) | 5.9 |
| Stearoyl-CoA Desaturase 1 (SCD1) | 4.0 |
| Stearoyl-CoA Desaturase 2 (SCD2) | 4.7 |
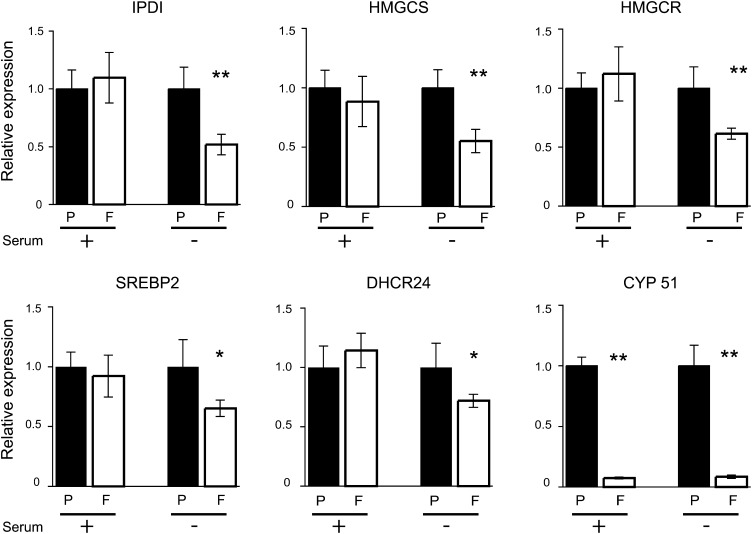
qPCR evaluation of mRNA expression from six genes involved in cholesterol biosynthesis (HMG-CoA reductase, HMG-CoA synthase, isopentenyl diphosphate isomerase, sterol regulatory protein 2, 24-dehydrocholesterol reductase, and lanosterol 14α demethylase) revealed decreased mRNA levels of approximately 40–50% in serum-starved cells except for lanosterol 14α demethylase (known as CYP51 and referred as such thereafter), which was repressed down to 5% expression levels compared with serum-fed and starved Puro controls (Fig. 2).
Fig. 2.
Expression of cholesterol biosynthesis genes by qPCR in Puro (P) or HA-Foxo 4 (F) 3T3L1 fibroblasts, serum fed (+) or serum starved (−). mRNA expression gene levels in HA-FoxO4 cells are displayed as normalized to those of Puro cells. Abbreviations of gene names are in the text. * and ** denote P values < 0.05 or < 0.01, respectively.
HA-FoxO4 inhibits cholesterol biosynthesis and induces the accumulation of DHL
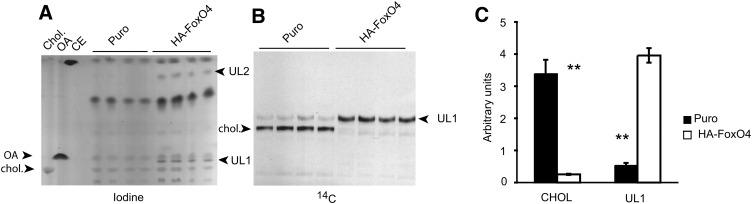
The effect of HA-FoxO4 on the disruption of cholesterol biosynthesis was unraveled by lipid analysis of Puro and HA-FoxO4-overexpressing cells. TLC of lipid extracts revealed that HA-FoxO4-overexpressing cells accumulated two characteristic unknown lipids, UL1 and UL2 (Fig. 3A). De novo lipid biosynthesis assessed by 14C-acetate incorporation revealed overwhelming inhibition of cholesterol biosynthesis and buildup of UL1 in HA-FoxO4 cells (Fig. 3B). Compared with Puro cells, cholesterol biosynthesis in HA-FoxO4 cells was reduced by 97% (P < 0.01), while UL1 was concomitantly increased by 7.3-fold (P < 0.01) (Fig. 3C).
Fig. 3.
Accumulation and characterization of an HA-FoxO4-specific lipid. A: TLC plate stained with iodine vapor showing fractionation of lipid extracts from Puro and HA-FoxO4 cells. Each lane is from a single well of a 6-well plate. HA-FoxO4 extracts show two additional unknown lipids, UL1 and UL2. B: TLC plate autoradiograph of lipid extracts from Puro and HA-FoxO4 cells showing de novo synthesis of 14C-labeled cholesterol (chol) in Puro cells and the 14C-unknown lipid (UL1) in HA-FoxO4 cells. C: Densitometric scan of cholesterol and the HA-FoxO4 lipid from the autoradiograph shown in B. **P < 0.01. Lipid markers are Cholesterol (chol), oleic acid (OA), and cholesteryl oleate, a cholesteryl ester (CE).
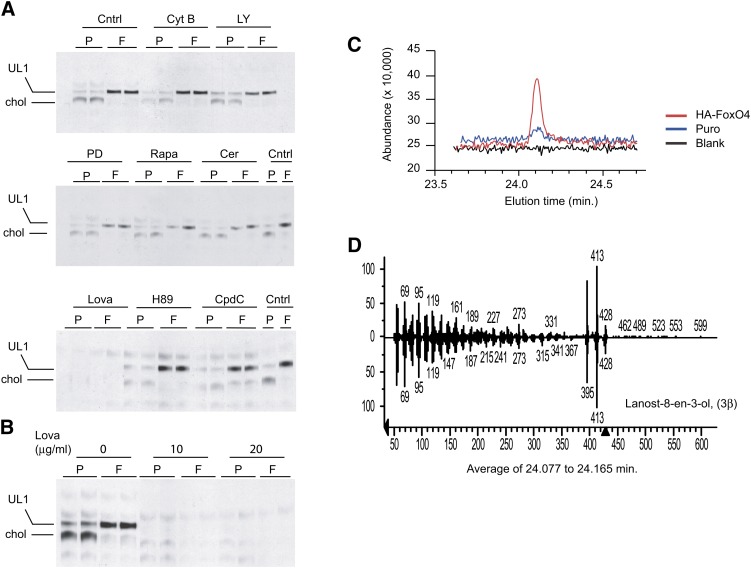
To uncover the nature of UL1, we treated Puro and HA-FoxO4 cells prior to 14C-acetate incorporation with inhibitors to various pathways, such as for glucose uptake (cytochalasin B), for phosphoinositide 3-kinase (LY 294002), for MAP kinase (PD9805), for mammalian target of rapamycin (rapamycin), for fatty acid biosynthesis (cerulenin), for cholesterol biosynthesis (lovastatin, an HMG-CoA reductase inhibitor), for protein kinase A (H89), and for AMP kinase (compound C). Lipids were then extracted and fractionated by TLC and the plates subjected to autoradiography. The resulting data demonstrate that only lovastatin (Fig. 4A, B) could inhibit the biosynthesis of cholesterol and of UL1, suggesting that UL1 consists of a sterol intermediate in the cholesterol biosynthesis pathway.
Fig. 4.
Characterization of the HA-FoxO4 UL1 lipid. A: TLC plate autoradiographs of lipid extracts from Puro (P) and HA-FoxO4 (F) fibroblasts either untreated (Cntrl) or pretreated with 10 μM cytochalasin B (Cyt B), 50 μM LY294002 (LY), 50 μM PD 98059 (PD), 0.1 μM rapamycin (Rapa), 10 μM cerulenin (Cer), 20 μg/ml lovastatin (Lova), 5 μM H89, or 20 μM compound C (CpdC) prior to 14C-acetate labeling. Lipids are labeled as cholesterol (chol) and the FoxO4 unknown lipid as UL1. Each lane represents a lipid extract from two combined wells of a 24-well plate. Inhibition of cholesterol and UL1 are seen with lovastatin treatment. B: TLC plate autoradiograph of lipid extracts from 14C-labeled Puro and HA-FoxO4 cells treated with two different concentrations of the HMG-CoA reductase inhibitor lovastatin (Lova). C: GC and elution times of blank, Puro, and HA-FoxO4 lipid extracts. D: MS fingerprint of UL1, which consists of lanost-8-en-3-ol (3β), commonly called DHL.
UL1 and UL2 were unveiled by GC-MS. The mass spectrum of purified UL1 revealed it to consist of DHL, a sterol intermediate in the late steps of cholesterol biosynthesis (Fig. 4C, D). UL2 consisted of TAGs containing predominantly palmitic acid and no evidence of either cholesterol or DHL (data not shown). These findings demonstrate that HA-FoxO4 overexpression induces the accumulation of the sterol DHL and of palmitate-rich TAGs.
FoxO4 induces the accumulation of lipid droplets, stimulates fatty acid biosynthesis, and enhances basal glucose uptake
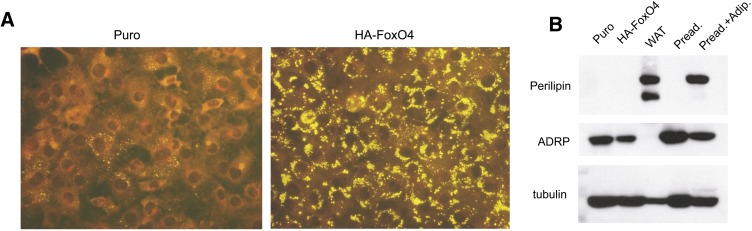
Growth of HA-FoxO4 cells to confluence in standard culture medium resulted in a noticeable increase of intracellular lipid droplets (Fig. 5A), raising the question whether HA-FoxO4 cells have switched, at least partly, to the adipogenesis program and started to accumulate neutral lipids. To address this question, we determined the nature of the proteins that coat lipid droplets in HA-FoxO4 cells. It is known that fibrobasts/preadipocytes and adipocytes coat their lipid droplets with the adipocyte differentiation-related protein (ADRP) and perilipin, respectively (15). Thus, the ADRP to perilipin switch serves as a marker for adipocyte differentiation. The nature of the proteins that coated the lipid droplets in HA-FoxO4 cells was identical to that in Puro cells and consisted of ADRP only, not the adipocyte-associated protein perilipin (Fig. 5B). Thus, lipid droplets in HA-FoxO4 cells retain their fibroblast/preadipocyte nature.
Fig. 5.
Accumulation of lipid droplets and determination of their protein coating in Puro and HA-FoxO4 cells. A: Nile red fluorescence staining of lipid droplets in HA-FoxO4 and Puro fibroblasts (200×). B: Western blots of protein lysates from fibroblasts (Puro and HA-FoxO4), white adipose tissue (WAT), 3T3L1 preadipocytes, and a mixture or 3T3L1 preadipocytes and adipocytes following induced differentiation. The Western blots were conjugated to antiperilipin, anti-adipocyte related protein (ADRP), or anti-tubulin (for protein loading).
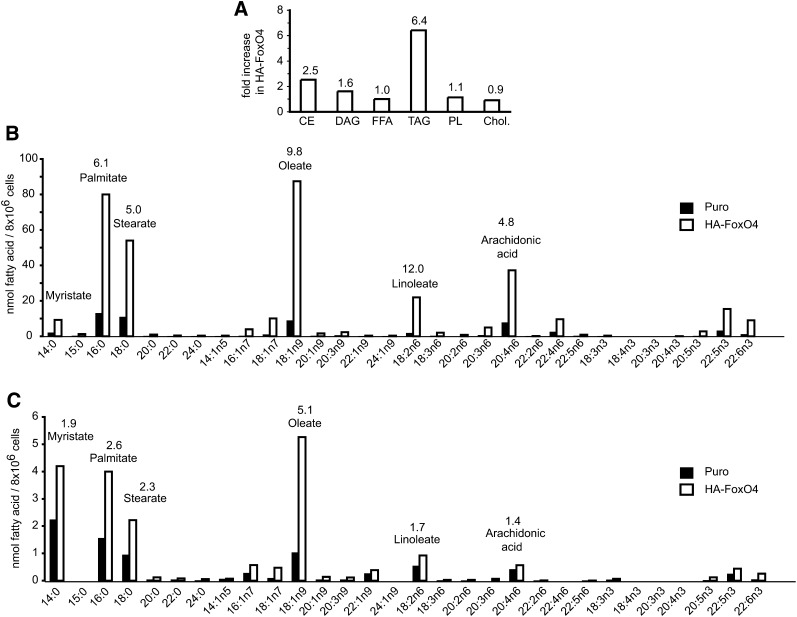
Lipid extracts from Puro and HA-FoxO4 cells were profiled for the nature of their fatty acids esterified either to glycerol (TAGs) or cholesterol (cholesteryl esters). Overall, HA-FoxO4 cells accumulated 6.4-fold more TAGs than Puro controls (Fig. 6A). While free fatty acids were not significantly elevated, fatty acids resulting from the hydrolysis of TAGs and cholesteryl esters revealed a large increase of saturated and unsaturated fatty acids in HA-FoxO4 versus Puro cells (Fig. 6B, C), suggesting that fatty acids biosynthesis is increased but rapidly esterified to TAGs and cholesteryl esters. Specifically, the most predominant fatty acids, oleate, palmitate, stearate, and arachidonic acid, were elevated 5–10-fold over those in Puro cells. Similarly, in cholesteryl esters, oleate, palmitate, stearate, and myristate were approximately 2.5-fold more elevated in HA-FoxO4 than in Puro cells.
Fig. 6.
Identification of lipid fractions and fatty acids in Puro and HA-FoxO4 fibroblasts by HPLC and GC flame ionization. A: Relative increases in HA-FoxO4 versus Puro fibroblasts of cholesteryl esters (CE), diacylglycerols (DAG), free fatty acids (FFA), TAGs, phospholipids (PL), and cholesterol. The numbers above each bar denote the fold elevation of each specific lipid fraction in HA-FoxO4 versus Puro cells. Identification of fatty acids in TAGs (B) and in cholesterol esters (C) from Puro and HA-FoxO4 fibroblasts. The numbers above the bars represent the fold increases of the corresponding fatty acid in HA-FoxO4 versus Puro fibroblasts. Fatty acids are denoted according to their standard nomenclature.
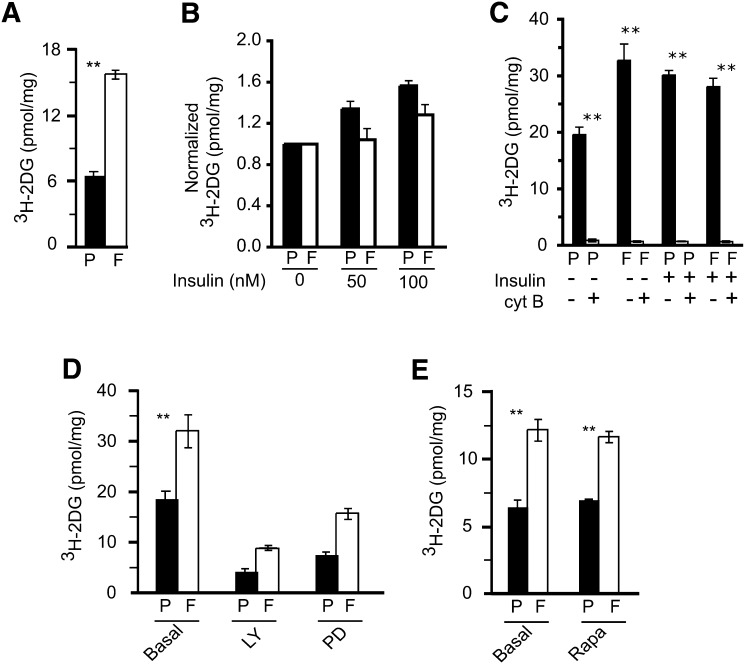
Another characteristic of HA-FoxO4 cells was a 2- to 3-fold increase in the uptake of 3H-2DG over Puro cells (Fig. 7A). Whereas 3H-2DG uptake was significantly increased in Puro cells upon stimulation with insulin, it was modestly but not significantly attenuated in similarly treated HA-FoxO4 cells (Fig. 7B). Treatment of basal- and insulin-stimulated Puro and HA-FoxO4 cells with cytochalasin B, a potent inhibitor of glucose transport, resulted in almost complete inhibition of 3H-2DG uptake in both cell types (Fig. 7C). Moreover, the glucose uptakes of Puro and HA-FoxO4 cells were sensitive to inhibitors targeting the phosphoinositide 3-kinase and MAP kinase but not to mammalian target of rapamycin (mTOR) (Fig. 7D, E). While these data show that basal glucose uptake is significantly increased in HA-FoxO4 cells, insulin stimulation and inhibitor studies suggest that the underlying mechanism of glucose uptake in HA-FoxO4 cells appears to be similar to that of Puro cells.
Fig. 7.
Uptake of 3H-2 deoxy glucose (3H-2DG) in Puro (P) and HA-FoxO4 (F) fibroblasts. Basal uptake (A) or pretreatments( B) with 50–100 nM insulin, 10 μM cytochalasin B (C), 50 μM LY294002 (LY) (D), 50 μM PD98059 rapamycin, or 1 μM rapamycin (Rapa) (E) prior to 3H-2DG uptake (n = 6 wells per group). Values are normalized to the protein content of each well. **P value < 0.01.
CYP51 inhibitors recapitulate the effects of FoxO4
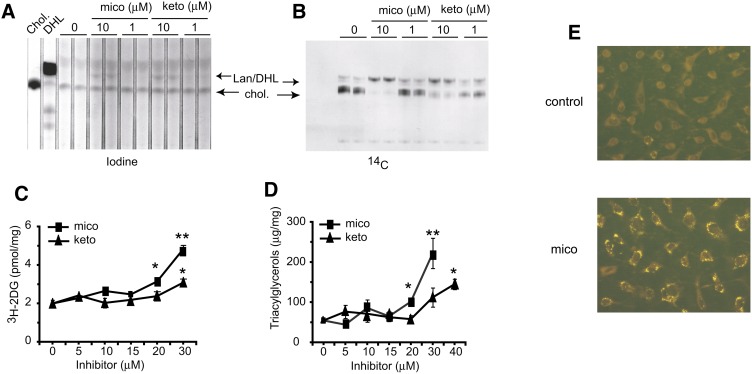
To reconcile the effects of HA-FoxO4 on glucose uptake and lipid droplet accumulation, we explored the possibility that these effects could be mediated by the inhibition of CYP51 independently of HA-FoxO4 overexpression. Having determined that lanosterol and DHL comigrate on the TLC plate and could thus not be independently resolved, we treated wild-type 3T3L1 fibroblasts with the CYP51 inhibitors, miconazole and ketoconazole. As expected, these treatments resulted in accumulation of lanosterol/DHL in steady-state cultures (Fig. 8A) and de novo biosynthesis (Fig. 8B). In addition, a dose-dependent increase in 3H-2DG uptake over basal untreated controls reached up to 2.4-fold (P < 0.01) with miconazole treatment and 1.5-fold (P < 0.05) with ketoconazole treatment (Fig. 8C). Furthermore, TAG accumulation was increased with both treatments. Miconazole induced TAG accumulation 1.9-fold (P < 0.05) and 4-fold (P < 0.01) over untreated control levels at 20 and 30 μM, respectively. Although ketoconazole was less effective than miconazole, it nevertheless induced at 40 μM a 2.7-fold increase (P = 0.02) in intracellular TAGs (Fig. 8D). Consistently, lipid droplets were readily increased in miconazole treated cells (Fig. 8E).
Fig. 8.
Effect of the CYP51 inhibitors, miconazole and ketoconazole, on cholesterol biosynthesis, 3H-2DG uptake, TAG synthesis, and lipid droplet accumulation in fibroblasts. Iodine vapor staining (A) and autoradiograph (B) of a TLC plate showing cholesterol (chol) and lanosterol/DHL (Lan/DHL) accumulation from lipid extracts of fibroblasts treated with vehicle or 1 or 10 μM miconazole or ketoconazole. C, D: Determination of 3H-2DG uptake and TAGs in fibroblasts treated with vehicle (0 μM) or increasing concentrations (5–40 μM) of miconazole (mico) or ketoconazole (keto) (n = 3 wells for each point on a curve). All determinations were normalized to the protein content of each well. *P < 0.05; **P < 0.01. E: Nile red fluorescence staining of lipid droplets in fibroblasts treated with vehicle control (0.1% DMSO) or 30 μM miconazole (200× magnification).
DHL but not lanosterol inhibits cholesterol biosynthesis in fibroblasts and hepatocytes
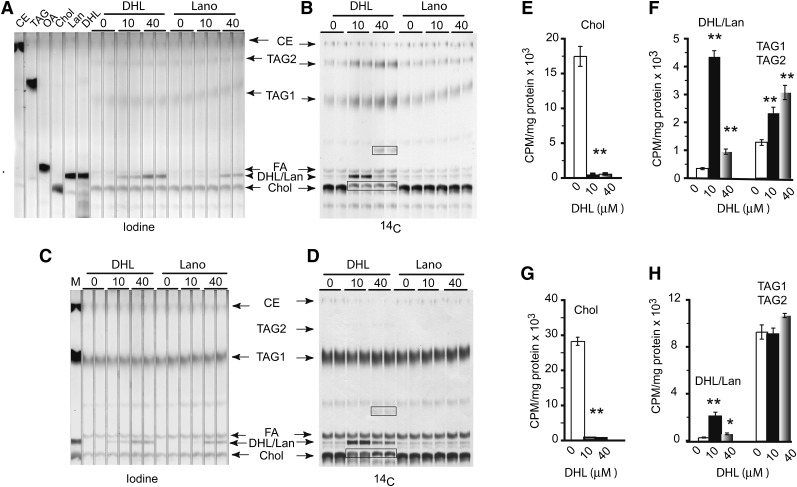
Since DHL and lanosterol could not be resolved by TLC fractionation, we addressed their respective effects on lipid biosynthesis in exogenously treated cells. The 3T3L1 fibroblasts took up either sterol in a dose-dependent fashion (Fig. 9A), and 14C-acetate incorporation revealed that only DHL and not lanosterol could inhibit de novo cholesterol biosynthesis at 10 or 40 μM (Fig. 9B), thus establishing the specificity of DHL on the inhibition of cholesterol biosynthesis. This finding was repeatedly determined with identical results, confirming the failure of lanosterol to inhibit cholesterol biosynthesis under the present conditions. In addition, treatment of Hepa 1-6 hepatocytes with DHL or lanosterol revealed the same findings, namely, that both sterols are taken up by cells in a dose-dependent fashion (Fig. 9C), but again only DHL inhibited de novo cholesterol biosynthesis, as noted by the absence of 14C-cholesterol in DHL-treated cells but its presence in lanosterol treated cells (Fig. 9D). It is also noticeable that in both fibroblasts and hepatocytes, the accumulation of 14C-DHL was more effective at 10 versus 40 μM, possibly due to the conversion of 14C-DHL/lanosterol to other unknown lipids at the higher exogenous DHL concentration (Fig. 9B, D).
Fig. 9.
Treatment of fibroblasts or hepatocytes with exogenous DHL or lanosterol. Iodine vapor stained lipid extracts from fibroblasts (A) or hepatocytes (C) treated with vehicle, DHL, or lanosterol (Lano) at 10 or 40 μM. B: Same TLC plate as in A. D: Same TLC plate as in C showing the de novo synthesis of 14C-labeled lipids. Note the accumulation of DHL or lanosterol in treated cells (as shown in iodine stained lipids) but only the inhibition of cholesterol biosynthesis in DHL-treated cells (as shown in 14C acetate labeling). Lipid markers are cholesterol oleate (CE), triolein (TAG), oleate (OA), cholesterol (Chol), lanosterol (Lano), and DHL. Identified lipid bands from extracts are cholesterol (chol), DHL or lanosterol (DHL/Lan), fatty acids (FA), TAGs (TAG1), palmitate-rich TAGs (TAG2), and two unknown lipids (denoted with boxes). Quantitation of de novo lipids biosynthesis in fibroblasts (E, F) and hepatocytes (G, H) (n = 6 per group). The radioactivity was normalized to the protein content of each lipid extract. *P value < 0.05; **P value < 0.01.
To quantitate relative biosynthesis levels of cholesterol, DHL, and TAGs following DHL treatment, we repeated the DHL treatments and recovered the radioactivity from each lipid band isolated from the respective TLC plates. Cholesterol was inhibited by 97–98% at both DHL concentrations in fibroblasts and hepatocytes (P < 0.01 each; Fig. 9E, G). Accumulation of DHL/lanosterol in 10 μM DHL-treated fibroblasts and hepatocytes was 12- and 7-fold higher than nontreated controls, respectively (P < 0.01 each). At 40 μM DHL, it was 2.1-fold (P < 0.01) and 2.7-fold (P < 0.01) in fibroblasts and hepatocytes, respectively (Fig. 9F, H). Moreover, in fibroblasts, TAGs were 1.8- and 2.4-fold increased at 10 and 40 μM DHL, respectively (P < 0.01; Fig. 9F). Identical treatment of hepatocytes failed to reveal significant changes in TAG accumulation, presumably due to their high endogenous levels of TAG biosynthesis (Fig. 9H). Overall, these experiments demonstrate the specificity of DHL toward the inhibition of cholesterol biosynthesis and the ability of DHL to divert lipid biosynthesis from sterol toward TAG accumulation.
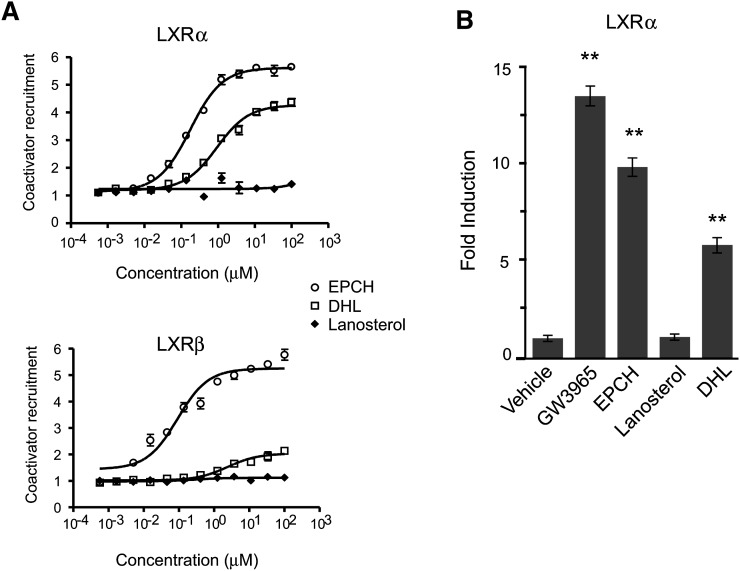
DHL is a strong activator of LXRα and binds to its ligand-binding domain
Because the effects of other sterols on gene expression are mediated by the nuclear receptor LXR (16), we sought to establish whether DHL could modulate the activity of this transcription factor. In a cell-free FRET-based coactivator recruitment assay, DHL showed an EC50 of approximately 1 μM for LXRα, demonstrating 5-fold less potency but similar efficacy to EPCH, the most potent natural agonist of LXRα and LXRβ. In contrast, lanosterol could not induce any LXRα coactivator recruitment activity and displayed a flat response. While EPCH showed an EC50 of ∼0.1 μM on LXRβ, DHL showed an EC50 of ∼5 μM, making it 50-fold less potent than EPCH and of marginal efficacy. Again, lanosterol was not an agonist of LXRβ (Fig. 10A). These data show that DHL is a strong direct agonist of LXRα and a weak activator of LXRβ, whereas lanosterol displays no activity on either LXRα or LXRβ. To examine whether DHL binding to LXRα has functional consequences, we also tested its ability to modulate LXRα activity in cells. In transient transfection assays, DHL stimulated the activity of a GAL-LXRα LBD reporter 5.8-fold over vehicle treated cells, while lanosterol had no activity. Compared with the agonist controls, DHL had 0.4- and 0.6-fold the activity of GW3965 or EPCH, respectively (Fig. 10B). Thus, DHL behaves as a functional ligand for LXRα.
Fig. 10.
DHL, but not lanosterol, is an LXR ligand. A: Coactivator recruitment assay of LXRα and LXRβ by FRET analysis. Dose-response curves of LXRα and LXRβ activation by DHL and lanosterol compared with EPCH. Data are means + SEM of two independent assays. B: The 10 T1/2 cells were transiently transfected with a GAL4-responsive luciferase reporter and an expression plasmid encoding a chimeric fusion of the GAL4 DNA binding domain and the LBD of hLXRα. Transfected cells were treated for 6 h with either the synthetic LXR ligand GW3965 (1 µM) or the sterols EPCH (9 µM), lanosterol (9 µM), or DHL (9 µM). Normalized luciferase values are expressed as fold induction versus DMSO; error bars indicate SDs. **P values < 0.01 compared with vehicle. All compounds tested except lanosterol activate GAL4-LXRα.
DISCUSSION
The various roles of the forkhead transcription factors in metabolism have emerged following the original finding that inactivation of the insulin receptor homolog daf-2 shifted the metabolism of C. elegans to fat storage, resulting in a prolonged life span (2). Since most of the metabolic effects of the FoxO transcription factors have been limited to FoxO1 (6, 7, 17), we undertook to investigate the sequelae of HA-FoxO4 stable overexpression in a newly derived preadipocyte cell line model where HA-FoxO4 is predominantly localized in the nucleus and thus presumably transcriptionally active. In this article, we report four findings: First, HA-FoxO4 inhibits cholesterol biosynthesis by repressing the expression of CYP51 and resulting in the accumulation of DHL. Second, DHL and not lanosterol inhibits cholesterol biosynthesis. Third, accumulation of DHL via HA-FoxO4 overexpression or pharmacologic inhibition of CYP51 enhances basal glucose uptake and TAG accumulation. Fourth, DHL is an activator of LXRα.
HA-FoxO4-mediated repression of CYP51 implicates FoxO4 in the regulation of the late steps of cholesterol biosynthesis, more specifically in the Kandutsch-Russell pathway (18), which consists of a series of demethylation and reduction steps of sterol intermediates leading to cholesterol. Although the mechanism by which HA-FoxO4 represses CYP51 expression is presently unknown, it could repress CYP51 promoter activity either directly or through a transcriptional complex that would otherwise activate CYP51 expression. Indeed, FoxOs were found to act as transcriptional corepressors (19) and to interact with Smad3 and Smad4 at the p21 promoter (20). Also, FoxO4 binds to and inactivates myocardin, which is a transcriptional activator of smooth muscle differentiation (21). The mechanism by which HA-FoxO4 selectively represses the CYP51 promoter and not other similarly induced SREBP2 promoters is likely to unveil a novel mode of regulating the late steps of cholesterol biosynthesis.
The inhibitory and detrimental effects of DHL accumulation on cholesterol biosynthesis suggest that intracellular DHL levels or those of any other sterol intermediates are critical and must be kept at low levels in order for cholesterol biosynthesis to proceed toward its final steps. Thus, the critical function of FoxO4 would be to regulate the CYP51 promoter, thereby preventing DHL levels from rising too high and inhibiting cholesterol biosynthesis. In this role, FoxO4 would be a major regulator of the Kandutsch-Russell pathway.
The accumulation of DHL, either as a result of HA-FoxO4-mediated inhibition of CYP51 expression or treatment with CYP51 inhibitors, clearly inhibits cholesterol biosynthesis. Interestingly, its closely related sterol lanosterol does not produce the same effect. Although this finding appears to contrast a previous report (22), which demonstrates that lanosterol stimulates the degradation of HMG-CoA reductase, it cannot be ruled out that the lanosterol-mediated degradation of HMG-CoA reductase is partially inhibitory to cholesterol biosynthesis. Considering that the only difference between lanosterol and DHL is the reduction of the C24-C25 double bond by Δ24 reductase in the sidearm chain of the steroid ring, this finding suggests that reduction of the C24-C25 double bond is essential for the inhibition of cholesterol biosynthesis. The finding that DHL and not lanosterol stimulates LXRα activity also implicates the C24-C25 region of the side chain as a critical component of the steroid ring.
The increase in basal glucose uptake by HA-FoxO4 cells coupled with their accumulation of DHL implicates LXR, namely, because activation of LXRα was shown to regulate GLUT4 expression and to improve glucose tolerance in mice fed a high-fat diet (23). Furthermore, other sterols besides DHL could stimulate LXRα downstream targets, such as the ABC transporters (24). Consistent with these findings, our observation that DHL binds to the ligand-binding domain of LXRα and strongly activates it suggests that LXRα could mediate the downstream effects of HA-FoxO4 on glucose uptake and lipid metabolism. Thus, enhanced glucose uptake will increase glucose oxidation and shuttle an additional supply of acetyl CoA to fatty acid biosynthesis, leading to intracellular TAG buildup, as presently determined for fibroblasts overexpressing HA-FoxO4.
Inhibition of CYP51 gene expression and resulting DHL accumulation could potentially be manipulated so that they could have a beneficial effect on insulin resistance and facilitate insulin-independent glucose uptake. The long-term extension of our studies suggests that the controlled accumulation of DHL could also be used to lower cholesterol biosynthesis and impact positively on disorders resulting from elevated cholesterol biosynthesis. Indeed, inhibition of cholesterol biosynthesis at the CYP51 step has been recognized and is consistent with previous efforts aimed at the derivation of a series of synthetic DHL analogs (25, 26) that target a decrease in HMG-CoA reductase activity. The ability of FoxO4 to inhibit cholesterol biosynthesis is likely to be most impacted in cells that are actively synthesizing cholesterol, such as in hepatocytes. In adipocytes, however, the efficiency of cholesterol biosynthesis is very low due to the accumulation of methylsterols (27, 28). Consistent with these observations was the lack of a detectable effect on cholesterol biosynthesis in transgenic mice overexpressing FoxO4 in adipocytes (12) (data not shown).
Overall, the cascade of events initiated by HA-FoxO4 on the repression of CYP51 gene expression uncovers a new role for FoxO4 in mediating the regulation of the late steps of cholesterol biosynthesis. Whether the action of FoxO4 is specific to CYP51 or could be extended to other promoters in the late steps of cholesterol biosynthesis remains to be determined; however, the interjection of FoxO4 in the regulation of the Kandutsch-Russell pathway is exciting and opens new avenues to investigate this often neglected arm of the cholesterol biosynthesis pathway.
Acknowledgments
We thank M. T. Burgering for providing the pBabe Puro and FoxO4 plasmids; Eric Yen and Robert Farese for advise on lipid analysis; Constantine Londos for the ADRP antibody; Divyan Patel and Bob Rooney at Genome Explorations (Memphis, TN) for microarray experiments and advice on pathway analysis; and Ruth Welti, Richard Jeanotte, and Devon L. Claycamp for GC-MS lipid analysis.
Footnotes
Abbreviations:
- ADRP
- adipocyte differentiation-related protein
- 2DG
- 2-deoxyglucose
- DHL
- 24,25 dihydrolanosterol
- EPCH
- 24(S),25 epoxycholesterol
- FRE
- forkhead-responsive element
- FRET
- fluorescence resonance energy transfer
- LBD
- ligand binding domain
- LXR
- liver X receptor α
- MAP
- mitogen-activated protein
- qPCR
- quantitative polymerase chain reaction
- TAG
- triacylglycerol
This work was supported by grants to F.F.C. from the UCSF Diabetes and Endocrinology Research Center (NIH P30-DK063720) and the UCSF Academic Senate. J.Z. was supported by National Institutes of Health training grant T32-DK07636. E.S. and N.M. were supported by National Institutes of Health Grant DK-081003 and by the McDonald's Center for Type 2 Diabetes and Obesity. The Kansas Lipidomics Research Center is supported by the National Science Foundation (EPS 0236913, MCB 0455318, and DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20RR16475), and the Functional Genomics Consortium Initiative of Kansas State University's Targeted Excellence program. The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Biggs W. H., 3rd, Cavenee W. K., Arden K. C. 2001. Identification and characterization of members of the FKHR (FOXO) subclass of winged-helix transcription factors in the mouse. Mamm. Genome. 12: 416–425. [DOI] [PubMed] [Google Scholar]
- 2.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature. 366: 461–464. [DOI] [PubMed] [Google Scholar]
- 3.Zhang W., Patil S., Chauhan B., Guo S., Powell D. R., Le J., Klotsas A., Matika R., Xiao X., Franks R., et al. 2006. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281: 10105–10117. [DOI] [PubMed] [Google Scholar]
- 4.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 389: 994–999. [DOI] [PubMed] [Google Scholar]
- 5.Medema R. H., Kops G. J., Bos J. L., Burgering B. M. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 404: 782–787. [DOI] [PubMed] [Google Scholar]
- 6.Nakae J., Kitamura T., Silver D. L., Accili D. 2001. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108: 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakae J., Kitamura T., Kitamura Y., Biggs W. H., 3rd, Arden K. C., Accili D. 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 4: 119–129. [DOI] [PubMed] [Google Scholar]
- 8.Furuyama T., Nakazawa T., Nakano I., Mori N. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosaka T., Biggs W. H., 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., Arden K. C. 2004. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA. 101: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Rajareddy S., Reddy P., Du C., Jagarlamudi K., Shen Y., Gunnarsson D., Selstam G., Boman K., Liu K. 2007. Infertility caused by retardation of follicular development in mice with oocyte-specific expression of Foxo3a. Development. 134: 199–209. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W., Cao Q., Peng Y., Zhang Q. J., Castrillon D. H., Depinho R. A., Liu Z. P. 2009. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. 137: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B., Zhu J., Mounzih K., Chehab E. F., Ke Y., Chehab F. F. 2009. Overexpression of the transcription factor Foxo4 is associated with rapid glucose clearance. Mol. Cell. Endocrinol. 307: 217–223. [DOI] [PubMed] [Google Scholar]
- 13.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 14.Molteni V., Li X., Nabakka J., Liang F., Wityak J., Koder A., Vargas L., Romeo R., Mitro N., Mak P. A., et al. 2007. N-Acylthiadiazolines, a new class of liver X receptor agonists with selectivity for LXRbeta. J. Med. Chem. 50: 4255–4259. [DOI] [PubMed] [Google Scholar]
- 15.Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. 1999. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10: 51–58. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin B. J., Zuercher W. J., Collins J. L. 2008. Recent advances in liver X receptor biology and chemistry. Curr. Top. Med. Chem. 8: 781–791. [DOI] [PubMed] [Google Scholar]
- 17.Altomonte J., Richter A., Harbaran S., Suriawinata J., Nakae J., Thung S. N., Meseck M., Accili D., Dong H. 2003. Inhibition of Foxo1 function is associated with improved fasting glycemia in diabetic mice. Am. J. Physiol. Endocrinol. Metab. 285: E718–E728. [DOI] [PubMed] [Google Scholar]
- 18.Kandutsch A. A., Russell A. E. 1960. Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J. Biol. Chem. 235: 2256–2261. [PubMed] [Google Scholar]
- 19.Ramaswamy S., Nakamura N., Sansal I., Bergeron L., Sellers W. R. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2: 81–91. [DOI] [PubMed] [Google Scholar]
- 20.Seoane J., Le H. V., Shen L., Anderson S. A., Massague J. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 117: 211–223. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. 2005. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell. 9: 261–270. [DOI] [PubMed] [Google Scholar]
- 22.Song B. L., Javitt N. B., DeBose-Boyd R. A. 2005. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1: 179–189. [DOI] [PubMed] [Google Scholar]
- 23.Laffitte B. A., Chao L. C., Li J., Walczak R., Hummasti S., Joseph S. B., Castrillo A., Wilpitz D. C., Mangelsdorf D. J., Collins J. L., et al. 2003. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc. Natl. Acad. Sci. USA. 100: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., et al. 2006. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281: 27816–27826. [DOI] [PubMed] [Google Scholar]
- 25.Trzaskos J. M., Fischer R. T., Ko S. S., Magolda R. L., Stam S., Johnson P., Gaylor J. L. 1995. Substrate-based inhibitors of lanosterol 14 alpha-methyl demethylase: II. Time-dependent enzyme inactivation by selected oxylanosterol analogs. Biochemistry. 34: 9677–9681. [DOI] [PubMed] [Google Scholar]
- 26.Trzaskos J. M., Ko S. S., Magolda R. L., Favata M. F., Fischer R. T., Stam S. H., Johnson P. R., Gaylor J. L. 1995. Substrate-based inhibitors of lanosterol 14 alpha-methyl demethylase: I. Assessment of inhibitor structure-activity relationship and cholesterol biosynthesis inhibition properties. Biochemistry. 34: 9670–9676. [DOI] [PubMed] [Google Scholar]
- 27.Durr I. F. 1966. The biosynthesis of squalene and sterols by the adipose tissue of rat, sheep and man. Biochem. J. 98: 317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tilvis R., Miettinen T. A. 1979. Effects of weight reduction on squalene, methyl sterols and cholesterol and on their synthesis in human adipose tissue. Eur. J. Clin. Invest. 9: 155–160. [DOI] [PubMed] [Google Scholar]