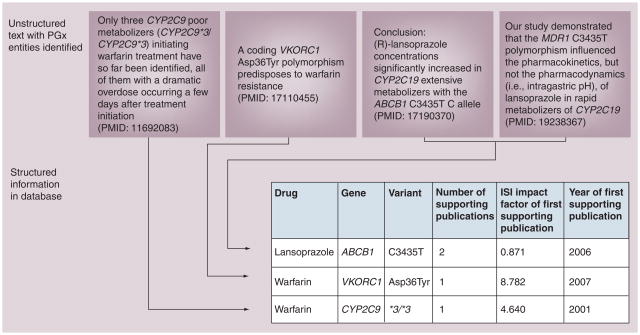
Figure 3. Information extraction: structuring unstructured data using text mining to create a database of facts.
Sentences appearing in publications are processed, key pharmacogenomics entities are identified and facts are used to populate a structured database. Note that MDR1 is a synonym for gene name ABCB1, a gene normalization system identifies this and resolves the issue. Multiple publications can support the same fact (e.g., lansoprazole–ABCB1–C3435T relationship). The database shown on the bottom contains ‘computable’ information: data structured in a table can easily be used and analyzed using software. Examples of tasks enabled by the information extraction include: identification of high-confidence relationships by mandating a minimum number of supporting articles per fact; identification of high-impact discoveries by mandating high-impact factor of the journal that was the first to publish the finding; identification of novel relationships by restricting ‘year of first supporting publication’ to the present year.
PGx: Pharmacogenomics.