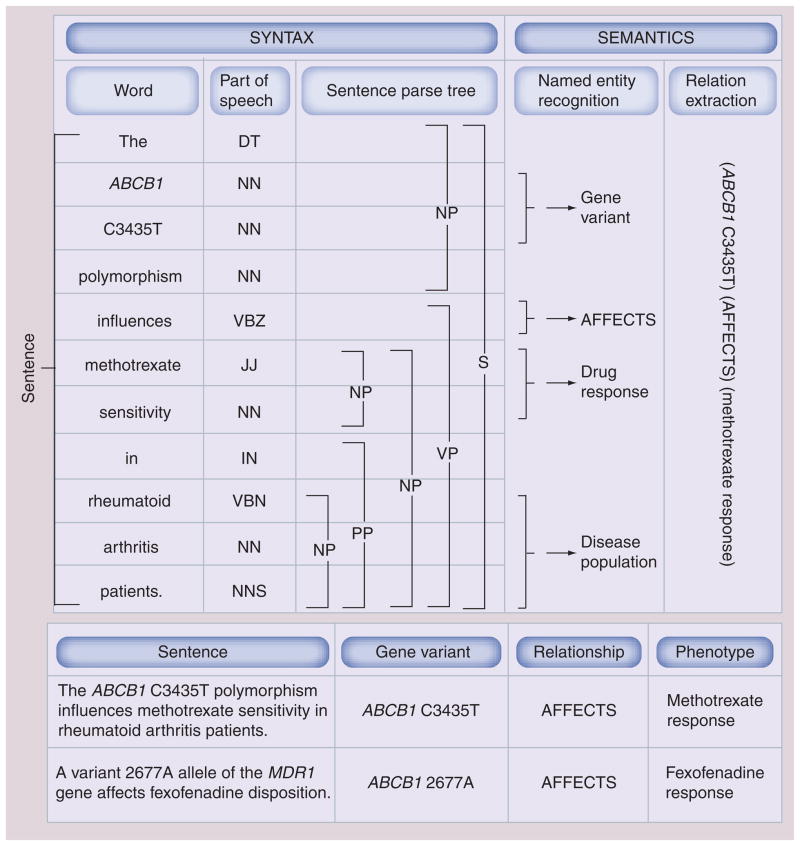
Figure 5. Levels of natural language processing analysis for a sentence.
The main levels of analysis are shown for the sentence ‘The ABCB1 C3435T polymorphism influences methotrexate sensitivity in rheumatoid arthritis patients.’ (PMID: 17181924) [137]. Processing uses syntax (sentence structure) and semantics (sentence meaning) to extract the relationship between gene variant and drug response. Sentence is tokenized into words, which are tagged with part of speech tags: DT, NNP, NN, VBZ, JJ, IN, NNS. Based on this sequence, parse tree is subsequently created for the sentence, which determines dependencies between words and groups words into phrases. Sentence parse tags: NP, PP, VP, S. Entities are recognized by combining the output of the syntactic analysis with external knowledge such as dictionaries of gene, drug and disease names in addition to categorization of relationship terms into classes (e.g., the class ‘AFFECTS’ would include the terms ‘affects’, ‘influences’, ‘has an effect on’; capitalization is used to indicate that this is a class name, not the textual term). Finally, relationship is extracted. Relationship term ‘influences’ found in raw text is normalized to <affects> class. Two rules are used: ‘Y sensitivity’ where Y is a drug, maps to Y <response>. Subsequently, the rule X <affects> Y <response> where X is a gene or protein or gene variant and Y is a drug is utilized. Using a similar process, the sentence ‘A variant 2677A allele of the MDR1 gene affects fexofenadine disposition.’ (PMID: 15536457) [138] can be processed to extract relationship between gene variant and drug response (‘MDR1’ maps to its synonym ABCB1, ‘disposition’ maps to <response>). Syntactic tagging is based on the output provided by the Stanford parser.
DT: Singular determiner; IN: Preposition; JJ: Adjective; NN: Singular or mass noun; NNP: Noun, singular proper; NNS: Plural noun; NP: Noun phrase; PP: Prepositional phrase; S: Sentence; VBZ: Verb, third person singular present; VP: Verb phrase.