Fig. 3.
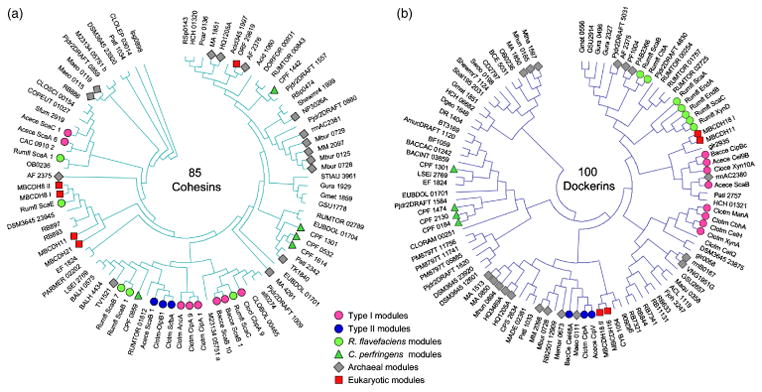
Phylogenetic distribution of representative cohesins (a) and dockerins (b) in the three domains of life. In the phylogenetic trees, 85 cohesin-like modules and 100 dockerin-like modules, derived from deduced amino-acid sequences of representative species, were aligned using the CLUSTALW program, which served to create an unrooted tree using the MEGA 4.1 program. Representative archaeal and eukaryotic cohesin and dockerin modules are labeled according to the designated key to facilitate comparison with those of representative cellulosomal modules (i.e. type-I, type-II, and Ruminococcus flavefaciens modules) and the recently described noncellulosomal modules of Clostridium perfringens; all other bacteria-derived modules are unlabeled on the phylogenetic trees. The terminology for cohesins and dockerins and the accession codes for their parent proteins are given in Tables S2 and S3, respectively.
