Figure 1.
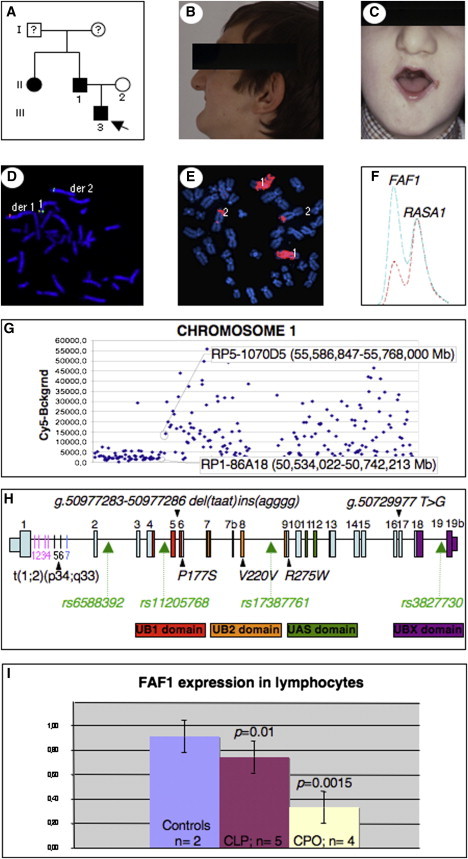
Identification of t(1;2) Breakpoint of the 46,XY,t(1;2)(p34;q33) Translocation
(A) Pedigree of family. An arrow indicates the proband; filled symbols indicate affected individuals.
(B and C) Father (during adolescence, after surgery), with micro/retrognathia, microstomia, and translocation.
(D) FISH with fosmid G248P84517F11 (green) spanning 1p breakpoint and fosmid G248P81885B3 (red) centromeric to 1p breakpoint.
(E) Reverse chromosome painting of microdissected derivative chromosome 1 red-labeled and hybridized to normal metaphases.
(F) MP/LC multiplex chromatogram for FAF1/RASA1. Chromatograms represent fluorescence intensity and are normalized to the RASA1 control amplicon. Blue indicates a control individual; red indicates a patient with a translocation. RASA1 is the Ras p21 protein activator 1 gene.
(G) Mapping of amplified microdissected aberrant chromosome 1 by array CGH. Dark squares indicate log-2-transformed intensity ratios. Clones are ordered from 1p tel to 1q tel. The first clones with decreased and increased intensities are indicated.
(H) FAF1 structure. Exons are positioned to scale. For colored exons, the FAF1 domains are denoted below. Probes 1–4 (pink bars in intron 1) were amplified and sequenced from aberrant chromosome 1; probes 5 and 6 (black) were not amplifiable from either derived chromosome, and probe 7 (blue) was amplified and sequenced from aberrant chromosome 2. Translocation breaks FAF1 in intron 1. Arrowheads indicate the position of identified changes and polymorphism; green arrows indicate SNPs genotyped in the Belgian cohort.
(I) qPCR of FAF1 on lymphocytes from controls, CL/P, and CPO patients.
