Abstract
Retroviruses integrate into the host cell’s chromosome. Accordingly, many aspects of the life cycle of retroviruses like HIV-1 are intimately linked to the functions of cellular proteins and RNAs. In this review, we discuss in brief recent genome-wide screens for the identification of cellular proteins that assist HIV-1 replication in human cells. We also review findings on other cellular moieties that help or restrict the viral life cycle.
Within the last ten years, assisted by new screening technologies for analysing cellular gene expression, it has become apparent that the interaction between HIV and its host cell is a vastly more sophisticated and intimate duet than had previously been surmised. Here we review the results of such approaches pertaining to HIV-1. Firstly we summarize the recent genome wide screens that have begun to reveal how extensively the virus interacts with and utilizes its host cell. We then survey the virus life cycle, highlighting the identified cellular chaperones of the virus and, where known, their role in viral replication. Finally we describe in some detail the functional attributes of certain of the best characterized innate defense molecules of the cell, the so called ‘Restriction factors’.
Genome wide evidence for positive and restrictive factors regulating HIV-1 replication
As in a number of other viral systems, such as influenza (1) and hepatitis C (2), the replication cycle of HIV-1 and its dependence on cellular factors has been studied by large-scale knockdown experiments using interfering RNA (siRNA and shRNA). These effectively identify factors necessary for replication and do not reveal cellular inhibitors of replication unless they are specifically designed to do so. Four such genome wide studies have been published. At first sight the two surprising features of these screens are the fact that so many cellular proteins are apparently involved in HIV-1 replication and secondly how little overlap there is between the factors identified in different screens. Some of the latter disparity may be ascribed to methodological differences in the cell lines, reporter genes, assay times and methods, as well as the nature of the infectious construct analyzed.
In the first of these screens, which assessed the whole viral life cycle, Brass et al. (3) transfected pools of siRNAs targeting over 20,000 host proteins into a HeLa derived cell line expressing both the cellular receptors for HIV-1 and an LTR driven reporter construct. Viral infection with the IIIB strain of HIV-1 followed 72 hours later. Virus production was analysed 48 hours later by direct fluorescent staining of the cells for capsid protein and by measuring infectivity of the supernatant from the cells. 273 cellular factors were identified as having an effect on HIV-1 of which 36 had previously been shown to play a role.
The second siRNA screen (4) targeted a similar number of proteins, focussing more on earlier stages of infection. In this case the 293T cells were infected at 48 hours with a pseudotyped HIV carrying the firefly luciferase gene within the viral RNA. Luciferase expression was a surrogate marker for successful infection, integration, and gene expression. From 295 initial hits, a combination of quantitative PCR and detailed in silico analyses reduced this to 40 proteins with possible roles in uncoating and reverse transcription and 15 more affecting integration or nuclear import.
The third screen (5), again using siRNA, targeted 20,000 gene products and, like the first of these screens, wild-type virus was used to infect a HeLa cell line expressing a reporter gene driven by an HIV-1 LTR. In this case wild type virus was used to infect the cells 24 hours after transfection, and the reporter gene assayed 24 and 72 hours later. siRNAs which affected cell viability were filtered out and potential targets reanalysed with a second siRNA pool. The final list of likely genes was arrived at by further in silico screening, limiting it to those genes expressed in activated T cells or macrophages. 311 targets resulted from this combined screen of which 44 had previously been implicated in HIV replication.
In total these screens identified 842 genes as capable of assisting HIV replication. This amounts to around 3.3% of the known human protein coding genes (6) The overlap however was small with 34 genes identified in two or more screens and only three, MED6, MED7 and RELA, common to all three (Fig. 1). Another surprise was the absence of some genes well-established as pivotal for HIV replication. LEDGF/p75, an essential cofactor for proviral integration (7) did not appear at all and TSG101 (8) and CRM1 (9), which are known to be essential, appeared in only one screen.
Figure 1.
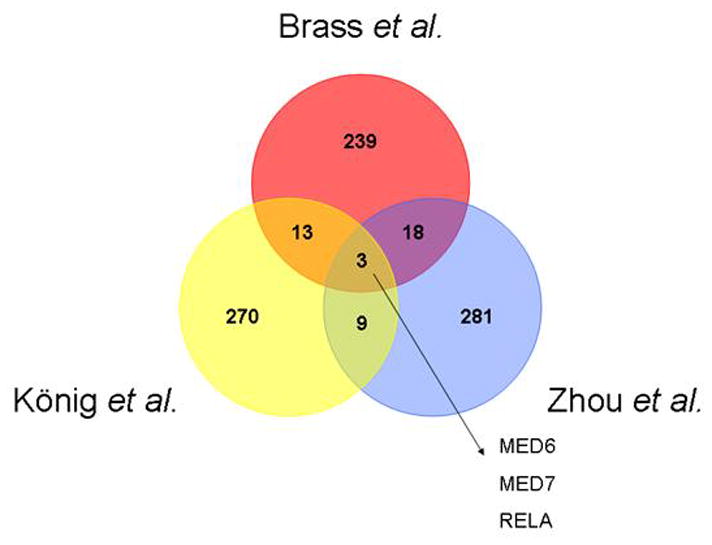
Diagram showing gene products unique, common to two, or common to all three siRNA screens (references 3, 4 and 5).
False negatives are not too surprising since in a high throughput screen there is no validation of successful knock down of the gene product. In addition the knock down may be only partial, the target may be essential for cell viability, or there may be functional redundancy, or a combination of these.
In addition complementarity of as little as seven nucleotides with an unrelated mRNA sequence may suffice for knockdown (10); hence, bystander interference with non-targeted genes, so-called off target effects, may also muddy the waters.
The cell line used must also be taken into account. So for example DDX53, identified in the Brass screen as affecting HIV replication is only expressed in testicular and some malignant tissues (11), making it an unlikely candidate HIV cofactor in vivo.
shRNA has also been used as a screen for cellular cofactors of HIV-1 (12) The fact that these are transcribed within the cell and processed to siRNA means that they have a more sustained duration of action. In a fourth genome wide screen, shRNAs that target 54,509 human transcripts were expressed as an FIV lentivirus vector library pseudotyped with VSV-G envelope and transduced into Jurkat cells. Antibiotic selection was used to pick out constitutively expressing cell clones. 9357 shRNA expressing clones survived selection, and these clones were then infected with HIV. Any cells surviving after four weeks of HIV infection were assumed to be expressing a shRNA that knocked down the expression of a protein necessary for viral replication. The findings from this study suggested that only 18.2% of the cell’s total transcriptome can be knocked down without affecting Jurkat cell viability in tissue culture implying that the durable knock down of 82% of cellular transcripts is incompatible with cell survival in culture. This screen identified 252 transcripts enriched in surviving cells and thus presumed to be required for HIV replication. Again there was trivial overlap with the three siRNA screens, with only three genes in common between this and each of the latter two, although combining all four screens revealed 40 genes common to at least two of them. The critical methodological difference is that the shRNA study incorporated a “selection” process built into the “screening” while the three siRNA studies are purely screening assays (Fig. 2).
Figure 2.
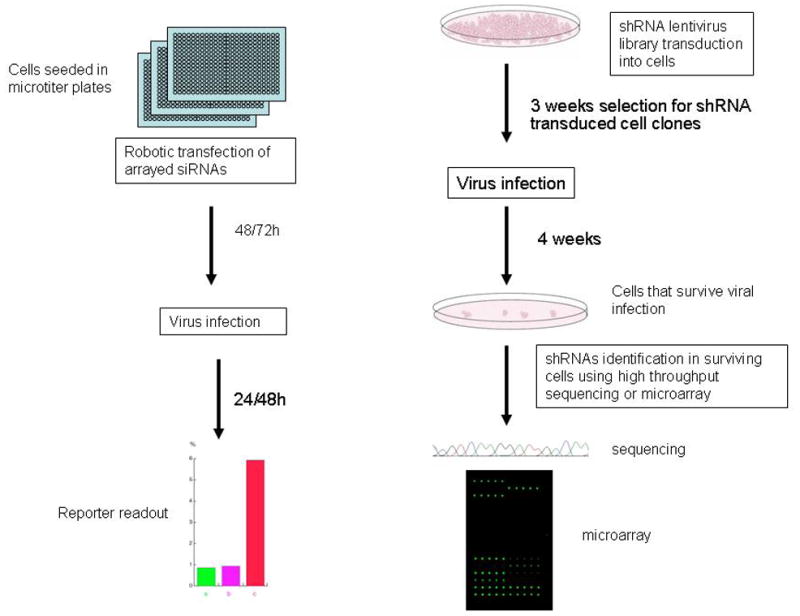
Schematic representations of general siRNA (left) and shRNA (right) based screening approaches for cellular factors that assist viral replication. More detailed explanation is in the text.
A comprehensive meta-analysis of the overlap between the three siRNA screens and other large-scale screens has been published. It concludes that potentially 2410 protein coding genes (9.5% all human genes) may be involved in the replication of HIV. The analyses also showed that variations between replicates, in time points, and in filtering thresholds all influence the readouts from the various siRNA screens. This article concludes that these type of approaches are better at identifying essential common cellular pathways rather than at pinpointing specific proteins within these pathways.
More recently a transcriptome analysis of unseparated cells from lymph nodes of HIV infected individuals has been published (13), seeking mRNAs which showed a correlation with viral load. The vast majority (~ 95% of 592 transcripts) showed a negative correlation. A significant number of those with identifiable function were involved in downregulating immune activation or cellular transcription, reflecting the enhancing effect on virus replication associated with activated immune cells. Surprisingly of the 5% (32 genes and 2 unknown transcripts) which were positively associated with viral load many were components of the innate and adaptive immune response, with a particular emphasis on the interferon pathway (which might be expected to inhibit HIV replication). Novel candidate restriction factors were identified. Perhaps because of the nature of the tissue of derivation being a mixed population of primary cells, there were only 5 genes identified which overlapped with the previous screens : GOLGA9P, MED31, TCFL5 (Cha), ACADSB, and CYCS
Known cellular factors involved in the HIV-1 lifecycle
In contrast to the above genome–wide screens there have been many studies focussing on particular stages of the viral life cycle and the cellular processes and individual cellular factors identified as being associated with each of these phases of HIV replication.
Viral entry
Surface receptor molecules for cell entry
The primary receptor for HIV, the CD4 protein, found on macrophages and T-lymphocytes (14) is a member of the immunoglobulin superfamily. The second major receptor (coreceptor) for HIV-1 is one of two molecules; either CCR5 on macrophages (15, 16) and T-lymphocytes, or CXCR4 found on T-lymphocytes (17). Both of these are chemokine receptors. CXCR4 is an alpha chemokine receptor specific for the ligand stromal derived factor 1 (SDF-1 or CXCL 12). CCR5 is a beta chemokine receptor, and like CXCR4 it belongs to the seven transmembrane family of cell surface receptors. Its ligands are a small family of molecules including MIP-1alpha (CCL3), MIP-1beta (CCL4) and RANTES (CCL5) (18) and variants including CCL3L1 and CCL4L1 (19). All of these ligands can compete with HIV and inhibit cell entry of the virus. The latter two have varying genetic copy numbers, and for CCL3L1 the number of gene copies and hence the level of chemokine was suggested to influence disease progression (20) although subsequent studies failed to substantiate this (21, 22).
In vivo chemokine receptors are signal transduction molecules, and there is evidence that this process is involved in HIV entry (23–25). There is accumulating evidence that infection involves disruption and rearrangement of the cytoskeletal molecules particularly actin. Downstream signalling from CXCR4, induced by HIV, dephosphorylates and activates cofilin leading to depolymerization of F-actin (26). Molecules inhibiting this process can impair HIV replication significantly. As yet a similar functional pathway has not been demonstrated for CCR5.
A more direct interaction has been implicated through the gp41 molecule directly recruiting p115-RhoGEF a guanine nucleotide exchange factor (27). This would facilitate remodelling of actin by GTPase activity.
Other cell surface molecules can also participate in HIV binding and entry. Integrin alpha4beta7 can bind gp120 activating LFA-1 which is involved in viral synapse formation and cell to cell transmission (28).
Transport of the viral preintegration complex to the nucleus
Subsequent to fusion and entry into the target cell, the steps required for the virus to traffic to the cell nucleus are relatively less well documented. Control of actin polymerization and depolymerization is apparently important, and the actin polymerization nucleator Arp2/3 is involved (29). It is suggested that this may facilitate a short range trafficking towards the microtubule network which is then responsible for transport to the nucleus. The actin cytoskeleton was also specifically assessed as playing a role in early HIV replication in one of the large siRNA screens. Proteins including AKAP13, another guanine exchange factor, were identified in this screen together with factors which regulate actin nucleation and organization as well as proteins affecting microtubule formation and function. Each of these candidates needs validation by specific studies.
Entry to the nucleus involves the ability to translocate across an intact nuclear membrane as, unlike other retrovirus families, lentiviruses can infect and integrate into non-mitotic cells. A number of the viral proteins which form the preintegration complex (PIC) have been shown to bind to members of the importin alpha family (30). They dock with Importin beta at the nuclear pore and facilitate entry of molecules bearing a nuclear localization signal. Several cellular proteins have been identified associated with the PIC including barrier to autointegration factor (BAF) (31, 32), Lap 2α (33) and HMG I(Y) protein (34). The viral accessory protein Vpr which is a component of the PIC binds to the nuclear pore complex protein Pom 121 (35). The PIC being larger than a nuclear pore, it is unsurprising that various other nuclear pore complex proteins have also been identified in nuclear entry including Nup 98 (36) Nup 124p (37), Nup 358 (38) and Nup 153 (39). Importin 7 has recently been shown to enhance nuclear entry of HIV-1 (but not HIV-2) (40) correlating with its ability to bind to the viral Integrase proteins which also are components of the PIC. Transportin 2 which was identified by two of the siRNA screens (3,4) has also been shown to enhance nuclear import of the PIC (41). Perhaps most intriguing has been the observation that tRNA molecules themselves can act as nuclear entry chaperones for the HIV PIC. Since most HIC cellular movement involves hijacking cellular processes it will be interesting to see how widespread this mode of nuclear targeting is (42).
Proteins influencing reverse transcription
AKAP14, a regulator of PKA in response to signal transduction from G protein coupled receptor, has been implicated in HIV infection, and there is evidence of direct interaction of this protein with the reverse transcriptase enzyme (43). Other proteins have proven to be more controversial. There are conflicting reports that the A/U binding protein HuR, known to stabilize mRNA, does, (44) and does not, (45) have a role in interacting with reverse transcriptase. Again the large scale siRNA screens have raised novel and plausible candidates including DHX15, a helicase, and RBM17, a nucleic acid binding protein, amongst others. Evidence of their direct interaction with the preintegration complex is awaited.
Transcription and chromatin
HIV-1 transcription is regulated by the viral promoter located in the 5′ long-terminal repeat (LTR) of the provirus. The LTR contains binding sites for several transcription factors such as Sp1 and NF-κB, NFAT, LEF-1, COUP-TF, Ets1, USF and AP-1. Amongst these factors Sp1 and NF-κB have been studied best, and through detailed mutagenesis of their binding sites in the LTR, their contributions to HIV-1 replication in human cells have been well-delineated (46, 47). Besides Sp1 and NF-κB, the roles of the other transcription factors are believed to contribute differentially to transcription under varying conditions of stimuli and in different cell types such as in primary T cells versus macrophages (48, 49). Because the activities of LTR-interacting DNA-binding factors in basal LTR-transcriptional initiation and elongation have been well-reviewed elsewhere (50, 51), they will not be further elaborated here.
In recent years, perhaps the biggest impetus for understanding the transcriptional regulation of HIV-1 arises from a need to address transcriptional mechanisms of proviral latency (52). Latently infected cells form a reservoir of antiretroviral treatment (ART) resistant cells that prevent curative therapy of HIV-1 (53, 54). These latent cells arise stochastically as a small population from productively HIV-1 infected cells that have integrated proviral DNA. To comprehend transcriptional latency, one needs to study the nucleosomally organized structure of the integrated provirus. HIV-1 integration is generally random but tends to favor active genes (55); however, independent of the site of integration in human chromosomes, two nucleosomes, named nuc-0 and nuc-1, are precisely organized in the 5′LTR. In particular, the histone organized nuc-1 structure (located at position −2 to +140 of the LTR) normally serves to down modulate basal transcription.
Because the nuc-1 nucleosome presents a barrier to HIV-1 transcription, it stands to reason that the HIV-1 encoded transcriptional activator Tat would have evolved mechanisms to resolve this block. Indeed, Tat is known to associate with histone acetyl transferase (HAT) proteins whose activities remodel nucleosomes to allow transcriptional access. Tat has been found to bind several different HATs: CBP/p300, p/CAF, GCN5, Tip60, and TAFII250 (56–60). Through binding to the HAT proteins, Tat then relieves chromatin repression at the HIV-1 LTR. Recently, Tat has also been found to bind a histone chaperone protein hNAP-1 (61) which acts with ATP-dependent chromatin remodeling complexes to facilitate transcription.
Countering the effect of HATs are the histone deacetylase proteins (HDAC) which remove the acetyl-group from HAT-acetylated histones to enforce transcriptional silencing. In the HIV-1 LTR, it is thought that the LSF protein binds position −10 to +27 of the LTR to recruit the YY1 factor which further binds HDAC-1 to silence viral transcription. Tat expression down regulates HDAC-1 serving to remove this repression of transcription. Indeed, this scheme of removal of repressive activity has been verified through treatment with several HDAC inhibitors (HDACIs) such as Trichostatin A (TSA), Trapoxin (TPX), Valproic Acid (VPA), sodium butyrate (NaBut) and other compounds which have been shown to activate integrated proviruses in latently infected cells (62, 63). The clinical importance of these findings lies in the potential use of HDACi in HIV-1-infected patients undergoing ART; this use could possibly activate the latent viral reservoirs allowing for the potential purging of the in vivo latently infected cells.
Post transcriptional regulation of incompletely spliced HIV-1 RNAs
The expression of unspliced and partially spliced HIV-1 RNAs is regulated post-transcriptionally by the viral Rev protein. Rev modulates the export of unspliced/partially spliced viral RNAs from the nucleus into the cytoplasm (64). This is an important property because unspliced and partially spliced viral RNAs serve as the moieties for the synthesis of Gag, Pol, and Env proteins; and the unspliced RNA is also the genomic RNA that is packaged into progeny virions. Because cellular RNAs are normally retained in the nucleus and do not exit into the cytoplasm, there must be a target-specificity by Rev for unspliced and partially spliced HIV-1 RNAs. This specificity is conferred on unspliced and partially spliced HIV-1 RNAs by a highly secondary structured RNA element (the Rev-responsive element, RRE) which is a binding site for the RNA-binding Rev protein. The current view is that Rev binds to the RRE and interacts with CRM1 (chromosome maintenance region 1 (65–67)) protein. This interaction then directs the viral ribonucleoprotein complex to a nuclear-cytoplasmic shuttling pathway which is normally used for the export of cellular small nuclear RNAs, and rRNAs. The RRE-CRM1 pathway is distinct from that used to export fully spliced HIV-1 mRNA and cellular mRNAs from the nucleus (68, 69). There are recent comprehensive reviews on the export of unspliced/ partially spliced HIV-1 RNA from the nucleus to the cytoplasm (70), on the possible role of Rev in the translation of HIV-1 transcripts (71), on Rev activity in RNA encapsidation (72), and on the effect of Rev on proviral integration (73). Rather than repeating those summaries, below, we will highlight selectively two classes of proteins, RNA helicases and RNA cap methylase, as examples of cellular factors that cooperate with Rev to regulate post transcriptional HIV-1 RNA expression.
It is perhaps not surprising that RNA helicases could serve as co-factors for Rev-directed export of HIV-1 RNAs (74). In this respect, DDX3 a cellular RNA helicase was found to enhance Rev-dependent nuclear-cytoplasmic export of HIV-1 RNAs (75). DDX3 is a nuclear-cytoplasmic shuttling protein which binds CRM1, Rev, and nuclear pore proteins. Thus, one notion is that this RNA helicase may function with Rev and CRM1 to remodel the HIV-1 ribonucleoprotein complex to ‘thread’ the attached RNA through the nuclear pore, facilitating its release into the cytoplasm. A second RNA helicase, DDX1, has also been reported to bind the N-terminus of Rev and to participate in the export of unspliced HIV-1 RNA from the nucleus to the cytoplasm (76). There is additional evidence that two other RNA helicases RHA and RH116 also regulate HIV-1 expression (77, 78). The mechanisms for these latter helicases in viral replication appear to be different from the nuclear-cytoplasmic regulation of RNA export. Knockdown of another helicase, DDX24, appears to reduce viral RNA encapsidation possibly by its negative effects on the enhancement of RNA packaging which is now a recognized property of the Rev protein. It is likely that additional RNA helicases will be discovered which interact with HIV-1.
Besides RNA-binding proteins, the inherent characteristics of an RNA may also dictate its post-transcriptional fate. An early RNA-modification of many cellular transcripts is the formation of a 7-methylguanosine (m7G) cap. The m7G cap facilitates the initiation of translation in mammalian cells, and uncapped RNAs are generally unstable (79, 80). The cap-status of HIV-1 RNAs had not been well-understood. Recently, it was found that different HIV-1 RNAs are either m7G- or hyper trimethylated TMG (trimethylguanosine) – capped at their 5′ ends (81). Viral transcripts containing RRE (i.e. unspliced or partially spliced HIV-1 RNAs) appear to be bound by Rev which then recruits a cap hypermethylating enzyme PIMT (peroxisome proliferator-activated receptor-interacting protein with methyltransferase) to modify the m7G-cap to a TMG-cap on these RNAs. The acquisition of a TMG-cap by these HIV-1 RNAs facilitates their recognition by CRM1 and directs the RNAs to the CRM1 nuclear-cytoplasmic export pathway. Accordingly, the PIMT-mediated TMG modification increases selectively the cytoplasmic expression and translation of HIV-1 mRNAs encoding for proteins like Gag and Env.
The above two classes of Rev-co-factors illustrate the complexities of post-transcriptional HIV-1 gene regulation. A potential benefit to characterizing these and other cellular cofactors for HIV-1 replication is that some of the proteins could be potentially targeted by small molecule inhibitors which might repress viral propagation. Initial candidate inhibitors for RNA helicases and cap hypermethylases and their possible utility for inhibiting HIV-1 have been reported (82).
Viral assembly and export
The intact viral particle consists of two copies of the virus RNA and a group of carefully ordered structural proteins. It appears that an early preassembly complex trafficking to the plasma membrane consists of a combination of a single RNA molecule with a small number of associated Gag proteins (83). The site of initial interaction of these two is not established although there is some evidence that this occurs at the microtubule organizing center (84, 85). Cellular proteins likely bind both the protein and RNA components of this complex to facilitate its trafficking. In the case of the RNA, a relatively small number of proteins have been implicated, some of which appear to be associated with the viral RNA in both nuclear and cytoplasmic subcellular compartments. Members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family including hnRNP which A1 and A2 possess nucleocytoplasmic shuttling capability have been identified as playing a role (86, 87). A1 appears to enhance Gag production possibly by increasing nuclear export of the genomic RNA although this is controversial. A2 binds to two response RNA elements within the genomic RNA, A2RE-1 and A2RE-2 found in the Gag and Vpr coding sequences respectively. Mutation of A2RE-2 leads to mislocalization of gRNA in and around the nucleus. Knockdown of hnRNPA −2 leads to accumulation of gRNA in the perinuclear microtubule organizing center (MTOC) however this RNA appeared to be derived from the cytoplasm. Conflicting results have been obtained when comparing knockdown of A2 with mutation of A2RE-A2, but with its suggested links to the microtubule system one could speculate that the protein is involved in ensuring the RNA takes the appropriate pathway as it traffics through the cell.
The RNA binding protein Staufen appears to act as a chaperone to the RNA and has been detected in viral particles (88). Similarities between this and the known HIV TAR RNA binding protein TRBP (89) may be pertinent.
There is increasing evidence that the microtubule network is involved in the cytoplasmic transport of the early assembly intermediates of HIV. Knockdown of KIF4, a kinin involved in cytokinesis which is also known to bind Gag altered localization of the latter, and expression of the dominant negative form of KIF4 decreased Gag levels globally but led to an accumulation in the perinuclear region. SOCS1 is induced during HIV infection and can stimulate late steps in HIV replication and can bind the MA and NC domains of Gag. Knockdown reduces trafficking and assembly and again results in the appearance of perinuclear aggregates of Gag. Other transport proteins including Arf and GGA may also play roles in trafficking and viral release (90).
Viral budding
In contrast to the relative sparsity of proteins known to be involved with trans-cytoplasmic trafficking, there is an abundance of information on those involved in the later stages of assembly and viral budding. This reflects the fact that the virus hijacks a complex of proteins within the cell which are usually used for budding and export into the endosomal system – the endosomal sorting complex required for transport (ESCRT) machinery. A full description and analysis of all of the ESCRT and ESCRT associated proteins involved in viral export are beyond the scope of this review. The reader is referred to excellent recent reviews on the subject (91–93). In brief it is known that the Gag protein of HIV can bind specifically to a number of the cellular proteins. In particular the PTAPP motif in the P6 region of Gag is able to interact directly with the ESCRT I protein TSG 101, and the YSPTL motif from P6 interacts with ALIX from ESCRT III. In a manner analogous to their role in budding of cell membranes into the endosome, these proteins facilitate the assembly of Gag monomers in an array at the plasma membrane and the evagination of a Gag containing particle away from the cell. The final process of membrane scission allowing the enveloped viral particle to escape from the cell surface is still a matter of debate but involves proteins of the CHMP family whose in vivo role is in the final separation of cell membranes during cell division.
Intracellular defences against HIV
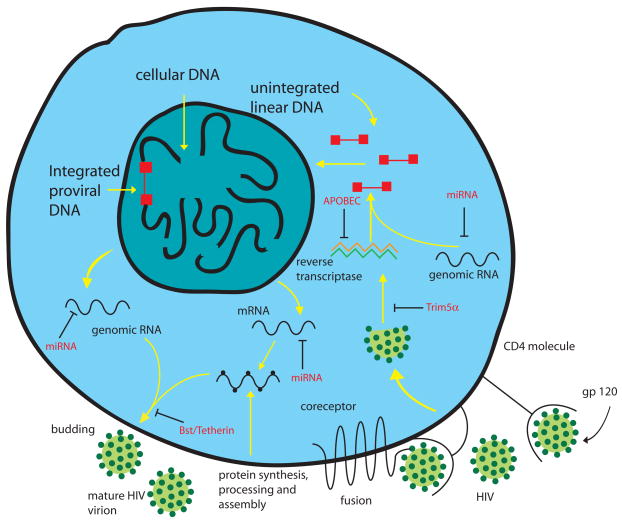
Clearly the cell is not a passive participant in virus replication. However apart from those multifarious pathways subverted by the virus for its own use there are inhibitory factors within cells which act as intracellular defences and whose presence inhibits or ‘restricts’ the virus (Fig. 3). The first of these to be identified in retroviruses was Fv1 which restricts ecotropic murine leukemia viruses (94). This paradigm prepared the way for the identification of similar factors restricting HIV. Because of their potential importance in novel antiviral approaches they have been extensively investigated in recent years.
Figure 3.
A drawing which highlights different infection processes used by HIV-1 and the various cellular factors that restrict viral replication. The drawing is modified after Wainberg and Jeang (216).
Trim 5 alpha
Simian immunodeficiency viruses are able to replicate in the cells of Old World monkeys (95–99), but HIV fails to do so (100) despite successful binding and cell entry. The inhibitory factor, initially identified and termed Lv1 (101–104) was saturable with an excess of viral cores. From a cDNA library of rhesus macaque expressed in HeLa cells, clones resistant to HIV-1 but sensitive to SIVmac were isolated and found to express simian TRIM5alpha (105) siRNA knockdown of this protein rendered the cells permissive to HIV. TRIM5alpha interacts with the viral capsid through a region also associated with the binding of a cellular factor Cyclophilin A (CypA) (106–108) a cellular peptidyl/prolyl isomerase which isomerises a peptide bond at this locus on the HIV-1 capsid. This was substantiated by identification of an unusual fusion protein in the owl monkey named TRIMCyp comprising a chimera of TRIM5alpha and cyclophilin (109, 110) (a similar insertion event producing a chimeric protein has been noted in monkeys of the Macaca genus (111). The owl monkey is a New World monkey yet still restricts HIV, and TRIMCyp was hypothesised to target the TRIM effector component to the viral core, since restriction could be blocked by the cyclophilin inhibitor Cyclosporin A.
TRIM stands for Tripartite Motif, a term used for a family of around 70 proteins sharing three polypeptide domains – an N-terminal RING domain, a B-box and a coiled coil. They carry out diverse functions within the cell including roles in development as well as anti-viral activity, and they have been implicated in oncogenesis (112–115). TRIM5alpha is the longest splice variant of the TRIM5 family with a unique C terminal B 30.2/SPRY domain. It is expressed constitutively but also upregulated by type I interferons (116–118). The RING domain has a zinc binding motif associated with E3 ubiquitin ligase activity, and TRIM5alpha can ubiquitinate other proteins and itself (119, 120). The B-box is a B2 form (121, 122) and is essential for the restriction activity of TRIM5alpha as shown by the inactivating effect of point mutations (123, 124). It also contains a zinc binding motif with homology to the RING domain although its function is still not fully elucidated.
The coiled coil domain, as occurs in other proteins such as the HIV envelope transmembrane glycoprotein, facilitates oligomerization, and the functional form of TRIM5alpha is thought to be either a dimer or a trimer (125).
The B30.2 domain contains a sequence with a PRYSPRY motif associated with exposed loops with three highly variable regions (V1-V3) (126, 127). These are thought to confer the virus binding specificity of the TRIM5alpha protein. As one of a number of examples, a single amino acid substitution in the V1 region substituting a non-positively charged amino acid for arginine at position 332 renders human TRIM5alpha as effective as owl monkey TRIMCyp in inhibiting HIV-1 (128, 129).
The exact mode of action of TRIM5alpha in restricting HIV-1 remains to be fully elucidated. Mutational analysis of the RING domain has produced inconsistent results. Ubiquitination of the viral capsid, followed by degradation in the proteasome, is suggested by the reduction in inhibition seen in the presence of proteasome inhibitors (130, 131), and it has been postulated that TRIM5alpha binds to capsid and that its own autoubiquitination capability flags the complex for degradation (132). However, other studies have suggested a RING dependent proteasome independent mechanism (133) possibly involving direct disassembly of the capsid.
Despite a high level of species specificity and diversity of TRIM5alpha between species and polymorphisms within a species the human sequence is almost invariant (134). This has been speculated to have contributed to the near universal susceptibility of humans to HIV (135). A small number of single nucleotide polymorphisms have been identified; the significance of these is unclear although one, the H43Y mutation, actually abolishes the moderate level of restriction achieved against HIV-2 (136, 137). Attempts to exploit this natural antiviral system to protect against HIV are being explored.
APOBEC
APOBEC3 proteins are a family of DNA editing proteins (138), named because of their homology to APOBEC1 an mRNA editing enzyme. There are seven in humans: - APOBEC 3A-3H which have arisen through gene duplication on chromosome 22. Overall this family of cytidine deaminases act to defend the cell against exogenous and endogenous retroelements and a variety of other viruses including HBV (139, 140) and HCV (141). APOBECs 3G and 3F are expressed in primary T-cell lines and in monocytes and macrophages and are the major ones involved in HIV restriction. They have target sequence preferences:- 3F targeting 5′-TC, whilst 3G is specific for 5′-CC, both deaminating the 3′ C residue. Their importance in HIV infection came to light during studies on the Vif protein, a 23kDa protein accessory protein of the virus which is required for replication in certain cell lines (142–144). APOBEC3G was found to be an endogenous restriction factor which could be overcome by the Vif protein (145). APOBEC3G is incorporated into the viral particle in the producer cell line and can exert its effect whether the virus subsequently infects APOBEC 3 expressing or non-expressing cells.
APOBEC 3F/G has cytidine deaminase activity and binds to the nucleocapsid protein and the viral genomic RNA (146) where, during the process of reverse transcription, it acts to deaminate cytidine to uridine on the negative strand of the viral cDNA as it is synthesized. The mutated cDNA now acts as template for the second DNA strand producing A to G mutations. Thus the provirus will contain multiple nonsense and stop codons and is nonfunctional. The mutated DNA will also be subject to destruction by the cell prior to integration (147–154).
APOBEC appears to have a number of other actions which are unrelated to the cytidine deaminase activity and active site mutants still display an inhibitory effect on HIV. The nature of this deaminase independent restriction is not fully elucidated (155–161). The deaminase activity is in the C-terminus of the protein, yet surprisingly mutants with an intact N terminal genomic RNA binding domain but a mutated C terminus can still cause A to G mutation.
Vif has been shown to target APOBEC3F/G to the proteasome through ubiquitination via linkage to the ELONGINB/C-CULLIN5 E3 ubiquitin ligase (162). There is also evidence that Vif can inhibit APOBEC translation, reducing levels in the producer cell (163, 164). It is highly species specific. A single point mutation at position 128 switches HIV-1 Vif restriction activity from HIV to SIVmac. (165–167). The concentration dependence of Vif activity and the selective advantage to HIV of mutational escape has fostered the concept that limited APOBEC activity may actually be advantageous to the virus in enhancing its own capabilities of sequence variation (168).
BST/Tetherin
The viral accessory protein Vpu had long been known to be essential for efficient virus assembly and export in some cell lines but not others (169–171). Mutating or deleting Vpu led, in non-permissive cells, to a phenotype of reduced viral production and accumulation of virus particles in the endosomes and at the cell surface (172). Initially Vpu was thought to interfere with an unwanted premature interaction of the virus surface (SU) protein gp120 with its cognate receptor CD4, as both were synthesised in the endoplasmic reticulum (173). However accumulating evidence showed that Vpu enhances virion release and overcomes a dominant but protease sensitive inhibitor which retains virions associated with the cell membrane (174, 175). Electron microscopy studies showed mature virions tethered to the plasma membrane (176). The cellular protein responsible for this restriction was only recently identified as CD317 or BST, also termed tetherin (177). Tetherin is a 30–36kDa heterogeneously glycosylated type II membrane protein which is an interferon inducible protein with an unusual topology. It has an N-terminal cytoplasmic tail, a transmembrane domain, an extracellular coiled coil domain and a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor. The intracellular domain of Tetherin contains a variety of important motifs; specifically a YxY domain mediating clathrin-linked endocytosis, a KxxK motif required for degradation by the KSHV K5 protein and, in non-human primates, a DDIWK sequence targeted by Nef and also resulting in degradation. The coiled-coil consists of two alpha helices containing three cysteines mediating disulphide bonding and two asparagines representing putative glycosylation sites in the ectodomain. Tetherin is enriched in lipid rafts of the plasma membrane due to its GPI anchor (178), where it can be incorporated into assembling virions and subsequently prevent their budding away from the cell surface. The mechanism is believed to involve bridging of the virion to the cell surface by the two membrane binding domains of the protein (179). The bridging complex appears to be an antiparallel dimer (180). Vpu counteracts the effect of tetherin by inducing its downregulation from the cell surface and its subsequent degradation (1). One model suggests an interaction between the two proteins’ transmembrane domains and subsequent ubiquitination of the tetherin moiety leading to proteasome degradation (181–183). Tetherin restricts the budding of a number of enveloped viruses, including a variety of retroviruses, Kaposi’s sarcoma herpesvirus (184)and Ebola virus (185). Vpu is species/virus specific.
MicroRNAs
Small non-coding RNAs play important roles in the regulation of mammalian genes. It has been suggested that over 30% of all human genes are regulated by microRNAs (miRNAs). To date, over 1,000 human miRNAs have been identified (www.microrna.org). While recent genome-wide siRNA and shRNA screenings (see above) have shown that several hundred host cell proteins contribute to the regulation of HIV-1 infection in human cells, how miRNA-mediated regulation complements this picture is poorly understood. The biogenesis and currently accepted mechanisms of action for miRNAs have been recently reviewed (186). We briefly outline below findings relevant to miRNA regulation of HIV-1.
Plants and lower eukaryotic cells use miRNAs as a form of RNA-interference (RNAi) to restrict infecting viruses (187). While mammals conserve the same functional miRNA repertoire and RNA-silencing machinery, some have debated whether they employ a miRNA-based antiviral strategy (12, 188–190). For endogenous mammalian retroviruses, there is a large body of literature demonstrating that a variety of small non-coding RNA forms are employed to silence these elements (191–193). In silico analyses have also indicated that exogenous mammalian viruses may be similarly susceptible to miRNA-based restriction (194, 195). Indeed, several investigators have demonstrated independently that a number of human miRNAs specifically influence productive HIV-1 infection in human cells (196–199). These results agree with findings that the knock down of either the Dicer protein (200) or the RISC components (201), both necessary for miRNA-mediated gene silencing, has resulted in enhanced HIV-1 replication in cells. The notion that miRNAs restrict viruses in mammals as they do in invertebrate or plant cells is additionally supported by increasing examples of RNAi-silencing suppressors encoded by mammalian viruses such as Adenovirus (202–204), HCV (205) Ebola (206) Influenza A virus (207–209), primate foamy virus (210), HIV (211–213) SARS corona virus(214) and HTLV-1 (215). Further investigation is needed to understand how RNA-based and protein-based viral restriction mechanisms cooperate together in human cells.
Concluding remarks
Our understanding of the extent of interaction and dependence of a virus like HIV-1 on cellular factors continues to increase as do the number of factors involved. Apart from giving us insights into the roles of these factors in the normal cell, they provide an array of novel drug targets. HIV-1 is such a mutable virus that drug treatments targeting pure virion processes rapidly produce escape mutations. Drugs that target processes which involve interactions with non-mutable cell proteins will also target regions of the virus whose variability is constrained by the conservation of their cellular partner. These present exciting new therapeutic opportunities in HIV-1 and other viruses.
Acknowledgments
AMLL is supported by grants from the Medical Research Council (UK) (G0801709, G0800142) and the Biomedical Research Centre (RG52162). KTJ is supported by intramural funds from NIAID, NIH, and by the Intramural Targeted Antiviral Program (IATAP) from the office of the Director, NIH.
Abbreviations
- HIV
human immunodeficiency virus
- siRNA
small interfering RNA
- shRNA
small hairpin RNA
References
- 1.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 4.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 8.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 10.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim WH, Yang H, Bang YJ, Jeoung DI. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem Biophys Res Commun. 2002;292:715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- 12.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT. Host genes associated with HIV-1 replication in lymphatic tissue. J Immunol. 2010;185:5417–5424. doi: 10.4049/jimmunol.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Blanpain C, Migeotte I, Lee B, Vakili J, Doranz BJ, Govaerts C, Vassart G, Doms RW, Parmentier M. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
- 19.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV’s switch-hitter. AIDS Res Hum Retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 21.Urban TJ, Weintrob AC, Fellay J, Colombo S, Shianna KV, Gumbs C, Rotger M, Pelak K, Dang KK, Detels R, Martinson JJ, O’Brien SJ, Letvin NL, McMichael AJ, Haynes BF, Carrington M, Telenti A, Michael NL, Goldstein DB. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15:1110–1112. doi: 10.1038/nm1009-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharya T, Stanton J, Kim EY, Kunstman KJ, Phair JP, Jacobson LP, Wolinsky SM. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15:1112–1115. doi: 10.1038/nm1009-1112. [DOI] [PubMed] [Google Scholar]
- 23.Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, Yoder A. Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 2009;5:e1000520. doi: 10.1371/journal.ppat.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, Liu J, Wang W, Vorster PJ, Agulto L, Stephany DA, Cooper JN, Marsh JW, Wu Y. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Yoder A, Yu D, Wang W, Liu J, Barrett T, Wheeler D, Schlauch K. Cofilin activation in peripheral CD4 T cells of HIV-1 infected patients: a pilot study. Retrovirology. 2008;5:95. doi: 10.1186/1742-4690-5-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fackler OT, Krausslich HG. Interactions of human retroviruses with the host cell cytoskeleton. Curr Opin Microbiol. 2006;9:409–415. doi: 10.1016/j.mib.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 29.Komano J, Miyauchi K, Matsuda Z, Yamamoto N. Inhibiting the Arp2/3 complex limits infection of both intracellular mature vaccinia virus and primate lentiviruses. Mol Biol Cell. 2004;15:5197–5207. doi: 10.1091/mbc.E04-04-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 31.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci U S A. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki Y, Yang H, Craigie R. LAP2alpha and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 2004;23:4670–4678. doi: 10.1038/sj.emboj.7600452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 35.Fouchier RA, Meyer BE, Simon JH, Fischer U, Albright AV, Gonzalez-Scarano F, Malim MH. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebina H, Aoki J, Hatta S, Yoshida T, Koyanagi Y. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 2004;6:715–724. doi: 10.1016/j.micinf.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Varadarajan P, Mahalingam S, Liu P, Ng SB, Gandotra S, Dorairajoo DS, Balasundaram D. The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol Biol Cell. 2005;16:1823–1838. doi: 10.1091/mbc.E04-07-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutten S, Walde S, Spillner C, Hauber J, Kehlenbach RH. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J Cell Sci. 2009;122:1100–1110. doi: 10.1242/jcs.040154. [DOI] [PubMed] [Google Scholar]
- 39.Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol. 2009;83:6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaitseva L, Cherepanov P, Leyens L, Wilson SJ, Rasaiyaah J, Fassati A. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology. 2009;6:11. doi: 10.1186/1742-4690-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 42.Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006;4:e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemay J, Maidou-Peindara P, Cancio R, Ennifar E, Coadou G, Maga G, Rain JC, Benarous R, Liu LX. AKAP149 binds to HIV-1 reverse transcriptase and is involved in the reverse transcription. J Mol Biol. 2008;383:783–796. doi: 10.1016/j.jmb.2008.08.055. [DOI] [PubMed] [Google Scholar]
- 44.Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain JC, Benarous R, Liu LX. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn J, Byeon IJ, Dharmasena S, Huber K, Concel J, Gronenborn AM, Sluis-Cremer N. The RNA binding protein HuR does not interact directly with HIV-1 reverse transcriptase and does not affect reverse transcription in vitro. Retrovirology. 2010;7:40. doi: 10.1186/1742-4690-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berkhout B, Jeang KT. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parrott C, Seidner T, Duh E, Leonard J, Theodore TS, Buckler-White A, Martin MA, Rabson AB. Variable role of the long terminal repeat Sp1-binding sites in human immunodeficiency virus replication in T lymphocytes. J Virol. 1991;65:1414–1419. doi: 10.1128/jvi.65.3.1414-1419.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology. 2009;6:118. doi: 10.1186/1742-4690-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colin L, Van Lint C. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology. 2009;6:111. doi: 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Nekhai S, Jeang KT. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 2006;1:417–426. doi: 10.2217/17460913.1.4.417. [DOI] [PubMed] [Google Scholar]
- 52.Mok HP, Lever AM. Chromatin, gene silencing and HIV latency. Genome Biol. 2007;8:228. doi: 10.1186/gb-2007-8-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson S, Reid AP, Kim S, Siliciano RF. Treatment implications of the latent reservoir for HIV-1. Adv Pharmacol. 2007;55:411–425. doi: 10.1016/S1054-3589(07)55012-X. [DOI] [PubMed] [Google Scholar]
- 54.Coleman CM, Wu L. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology. 2009;6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Col E, Gilquin B, Caron C, Khochbin S. Tat-controlled protein acetylation. J Biol Chem. 2002;277:37955–37960. doi: 10.1074/jbc.M206694200. [DOI] [PubMed] [Google Scholar]
- 57.Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J Biol Chem. 2001;276:28179–28184. doi: 10.1074/jbc.M101385200. [DOI] [PubMed] [Google Scholar]
- 58.Weissman JD, Brown JA, Howcroft TK, Hwang J, Chawla A, Roche PA, Schiltz L, Nakatani Y, Singer DS. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci U S A. 1998;95:11601–11606. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 60.Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- 61.Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology. 2008;5:8. doi: 10.1186/1742-4690-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Margolis DM. Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep. 2010;7:37–43. doi: 10.1007/s11904-009-0033-9. [DOI] [PubMed] [Google Scholar]
- 63.Savarino A, Mai A, Norelli S, El Daker S, Valente S, Rotili D, Altucci L, Palamara AT, Garaci E. “Shock and kill” effects of class I-selective histone deacetylase inhibitors in combination with the glutathione synthesis inhibitor buthionine sulfoximine in cell line models for HIV-1 quiescence. Retrovirology. 2009;6:52. doi: 10.1186/1742-4690-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris ME, Hope TJ. RNA export: insights from viral models. Essays Biochem. 2000;36:115–127. doi: 10.1042/bse0360115. [DOI] [PubMed] [Google Scholar]
- 65.Bogerd HP, Echarri A, Ross TM, Cullen BR. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otero GC, Harris ME, Donello JE, Hope TJ. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 68.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjold ML. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clouse KN, Luo MJ, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol. 2001;3:97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- 70.McLaren M, Marsh K, Cochrane A. Modulating HIV-1 RNA processing and utilization. Front Biosci. 2008;13:5693–5707. doi: 10.2741/3110. [DOI] [PubMed] [Google Scholar]
- 71.Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groom HC, Anderson EC, Lever AM. Rev: beyond nuclear export. J Gen Virol. 2009;90:1303–1318. doi: 10.1099/vir.0.011460-0. [DOI] [PubMed] [Google Scholar]
- 73.Grewe B, Uberla K. The human immunodeficiency virus type 1 Rev protein: menage a trois during the early phase of the lentiviral replication cycle. J Gen Virol. 2010;91:1893–1897. doi: 10.1099/vir.0.022509-0. [DOI] [PubMed] [Google Scholar]
- 74.Jeang KT, Yedavalli V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006;34:4198–4205. doi: 10.1093/nar/gkl398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 76.Fang J, Kubota S, Yang B, Zhou N, Zhang H, Godbout R, Pomerantz RJ. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 77.Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cocude C, Truong MJ, Billaut-Mulot O, Delsart V, Darcissac E, Capron A, Mouton Y, Bahr GM. A novel cellular RNA helicase, RH116, differentially regulates cell growth, programmed cell death and human immunodeficiency virus type 1 replication. J Gen Virol. 2003;84:3215–3225. doi: 10.1099/vir.0.19300-0. [DOI] [PubMed] [Google Scholar]
- 79.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 80.Schwer B, Mao X, Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 1998;26:2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yedavalli VS, Jeang KT. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yedavalli VS, Zhang N, Cai H, Zhang P, Starost MF, Hosmane RS, Jeang KT. Ring expanded nucleoside analogues inhibit RNA helicase and intracellular human immunodeficiency virus type 1 replication. J Med Chem. 2008;51:5043–5051. doi: 10.1021/jm800332m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 85.Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 2006;7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 86.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beriault V, Clement JF, Levesque K, Lebel C, Yong X, Chabot B, Cohen EA, Cochrane AW, Rigby WF, Mouland AJ. A late role for the association of hnRNP A2 with the HIV-1 hnRNP A2 response elements in genomic RNA, Gag, and Vpr localization. J Biol Chem. 2004;279:44141–44153. doi: 10.1074/jbc.M404691200. [DOI] [PubMed] [Google Scholar]
- 88.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Joshi A, Garg H, Nagashima K, Bonifacino JS, Freed EO. GGA and Arf proteins modulate retrovirus assembly and release. Mol Cell. 2008;30:227–238. doi: 10.1016/j.molcel.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 92.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Benaroch P, Billard E, Gaudin R, Schindler M, Jouve M. HIV-1 assembly in macrophages. Retrovirology. 2010;7:29. doi: 10.1186/1742-4690-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 95.Alter HJ, Eichberg JW, Masur H, Saxinger WC, Gallo R, Macher AM, Lane HC, Fauci AS. Transmission of HTLV-III infection from human plasma to chimpanzees: an animal model for AIDS. Science. 1984;226:549–552. doi: 10.1126/science.6093251. [DOI] [PubMed] [Google Scholar]
- 96.Gardner MB, Luciw PA. Animal models of AIDS. FASEB J. 1989;3:2593–2606. doi: 10.1096/fasebj.3.14.2556312. [DOI] [PubMed] [Google Scholar]
- 97.Himathongkham S, Luciw PA. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 98.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring Tlymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. Early replication block of human immunodeficiency virus type 1 in monkey cells. J Gen Virol. 1995;76(Pt 11):2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- 101.Besnier C, Takeuchi Y, Towers G. Restriction of lentivirus in monkeys. Proc Natl Acad Sci U S A. 2002;99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 106.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 107.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 108.Kootstra NA, Munk C, Tonnu N, Landau NR, Verma IM. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc Natl Acad Sci U S A. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 110.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci U S A. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao CH, Kuang YQ, Liu HL, Zheng YT, Su B. A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS. 2007;21(Suppl 8):S19–26. doi: 10.1097/01.aids.0000304692.09143.1b. [DOI] [PubMed] [Google Scholar]
- 112.Yoshigai E, Kawamura S, Kuhara S, Tashiro K. Trim36/Haprin plays a critical role in the arrangement of somites during Xenopus embryogenesis. Biochem Biophys Res Commun. 2009;378:428–432. doi: 10.1016/j.bbrc.2008.11.069. [DOI] [PubMed] [Google Scholar]
- 113.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y, Li Y, Qi X, Yuan W, Ai J, Zhu C, Cao L, Yang H, Liu F, Wu X, Liu M. TRIM45, a novel human RBCC/TRIM protein, inhibits transcriptional activities of ElK-1 and AP-1. Biochem Biophys Res Commun. 2004;323:9–16. doi: 10.1016/j.bbrc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 115.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Asaoka K, Ikeda K, Hishinuma T, Horie-Inoue K, Takeda S, Inoue S. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem Biophys Res Commun. 2005;338:1950–1956. doi: 10.1016/j.bbrc.2005.10.173. [DOI] [PubMed] [Google Scholar]
- 117.Carthagena L, Parise MC, Ringeard M, Chelbi-Alix MK, Hazan U, Nisole S. Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology. 2008;5:59. doi: 10.1186/1742-4690-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sakuma R, Mael AA, Ikeda Y. Alpha interferon enhances TRIM5alphamediated antiviral activities in human and rhesus monkey cells. J Virol. 2007;81:10201–10206. doi: 10.1128/JVI.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, Kaul S, Meroni G, Jensen JP, Weissman AM, D’Arpa P. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 120.Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 121.Torok M, Etkin LD. Two B or not two B? Overview of the rapidly expanding Bbox family of proteins. Differentiation. 2001;67:63–71. doi: 10.1046/j.1432-0436.2001.067003063.x. [DOI] [PubMed] [Google Scholar]
- 122.Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 123.Li X, Li Y, Stremlau M, Yuan W, Song B, Perron M, Sodroski J. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains. J Virol. 2006;80:6198–6206. doi: 10.1128/JVI.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Javanbakht H, An P, Gold B, Petersen DC, O’Huigin C, Nelson GW, O’Brien SJ, Kirk GD, Detels R, Buchbinder S, Donfield S, Shulenin S, Song B, Perron MJ, Stremlau M, Sodroski J, Dean M, Winkler C. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 126.Masters SL, Yao S, Willson TA, Zhang JG, Palmer KR, Smith BJ, Babon JJ, Nicola NA, Norton RS, Nicholson SE. The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues. Nat Struct Mol Biol. 2006;13:77–84. doi: 10.1038/nsmb1034. [DOI] [PubMed] [Google Scholar]
- 127.Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 129.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rold CJ, Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maegawa H, Miyamoto T, Sakuragi J, Shioda T, Nakayama EE. Contribution of RING domain to retrovirus restriction by TRIM5alpha depends on combination of host and virus. Virology. 2010;399:212–220. doi: 10.1016/j.virol.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 134.Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 135.Newman RM, Johnson WE. A brief history of TRIM5alpha. AIDS Rev. 2007;9:114–125. [PubMed] [Google Scholar]
- 136.Song H, Nakayama EE, Yokoyama M, Sato H, Levy JA, Shioda T. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J Virol. 2007;81:7280–7285. doi: 10.1128/JVI.00406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nakayama EE, Shioda T. Anti-retroviral activity of TRIM5 alpha. Rev Med Virol. 2010;20:77–92. doi: 10.1002/rmv.637. [DOI] [PubMed] [Google Scholar]
- 138.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O’Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 140.Rosler C, Kock J, Kann M, Malim MH, Blum HE, Baumert TF, von Weizsacker F. APOBEC-mediated interference with hepadnavirus production. Hepatology. 2005;42:301–309. doi: 10.1002/hep.20801. [DOI] [PubMed] [Google Scholar]
- 141.Chen WX, Shan LN, Chen J, Zhang ZZ, Zhang BQ, Huang AL. Inhibition of HCV IRES controlled reporter gene expression by RNA interference. Zhonghua Gan Zang Bing Za Zhi. 2006;14:521–524. [PubMed] [Google Scholar]
- 142.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 143.Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gabuzda DH, Li H, Lawrence K, Vasir BS, Crawford K, Langhoff E. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J Acquir Immune Defic Syndr. 1994;7:908–915. [PubMed] [Google Scholar]
- 145.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 146.Strebel K, Khan MA. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 148.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 149.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 151.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 152.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 153.Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Turelli P, Liagre-Quazzola A, Mangeat B, Verp S, Jost S, Trono D. APOBEC3-independent interferon-induced viral clearance in hepatitis B virus transgenic mice. J Virol. 2008;82:6585–6590. doi: 10.1128/JVI.00216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 157.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 158.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 159.Bonvin M, Greeve J. Hepatitis B: modern concepts in pathogenesis--APOBEC3 cytidine deaminases as effectors in innate immunity against the hepatitis B virus. Curr Opin Infect Dis. 2008;21:298–303. doi: 10.1097/QCO.0b013e3282fe1bb2. [DOI] [PubMed] [Google Scholar]
- 160.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 161.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 164.Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 167.Schrofelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 169.Yasuda Y, Miyake S, Kato S, Kita M, Kishida T, Kimura T, Ikuta K. Interferon-alpha treatment leads to accumulation of virus particles on the surface of cells persistently infected with the human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1990;3:1046–1051. [PubMed] [Google Scholar]
- 170.Dianzani F, Castilletti C, Gentile M, Gelderblom HR, Frezza F, Capobianchi MR. Effects of IFN alpha on late stages of HIV-1 replication cycle. Biochimie. 1998;80:745–754. doi: 10.1016/s0300-9084(99)80028-5. [DOI] [PubMed] [Google Scholar]
- 171.Sakai H, Tokunaga K, Kawamura M, Adachi A. Function of human immunodeficiency virus type 1 Vpu protein in various cell types. J Gen Virol. 1995;76(Pt 11):2717–2722. doi: 10.1099/0022-1317-76-11-2717. [DOI] [PubMed] [Google Scholar]
- 172.Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Terwilliger EF, Cohen EA, Lu YC, Sodroski JG, Haseltine WA. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci U S A. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gottlinger HG, Dorfman T, Cohen EA, Haseltine WA. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci U S A. 1993;90:7381–7385. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog. 2010;6:e1000701. doi: 10.1371/journal.ppat.1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 178.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell. 2010;102:335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Andrew AJ, Miyagi E, Kao S, Strebel K. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology. 2009;6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2009;83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 187.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Berkhout B, Jeang KT. RISCy business: MicroRNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 189.Grassmann R, Jeang KT. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.ter Brake O, Haasnoot J, Kurreck J, Berkhout B. ESF-EMBO symposium: antiviral applications of RNA interference. Retrovirology. 2008;5:81. doi: 10.1186/1742-4690-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 192.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 195.Watanabe T, Umehara T, Kohara M. Therapeutic application of RNA interference for hepatitis C virus. Adv Drug Deliv Rev. 2007;59:1263–1276. doi: 10.1016/j.addr.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 196.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 197.Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, Brahmachari SK. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 201.Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, Jacquet JM, Reynes J, Levy Y, Saib A, Bennasser Y, Benkirane M. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAIIderived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 206.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 209.de Vries W, Haasnoot J, Fouchier R, de Haan P, Berkhout B. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J Gen Virol. 2009;90:1916–1922. doi: 10.1099/vir.0.008284-0. [DOI] [PubMed] [Google Scholar]
- 210.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 211.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 212.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 213.Qian S, Zhong X, Yu L, Ding B, de Haan P, Boris-Lawrie K. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc Natl Acad Sci U S A. 2009;106:605–610. doi: 10.1073/pnas.0806822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Karjee S, Minhas A, Sood V, Ponia SS, Banerjea AC, Chow VT, Mukherjee SK, Lal SK. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J Virol. 2010;84:10395–10401. doi: 10.1128/JVI.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Abe M, Suzuki H, Nishitsuji H, Shida H, Takaku H. Interaction of human Tcell lymphotropic virus type I Rex protein with Dicer suppresses RNAi silencing. FEBS Lett. 2010;584:4313–4318. doi: 10.1016/j.febslet.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 216.Wainberg MA, Jeang KT. 25 years of HIV-1 research - progress and perspectives. BMC Med. 2008;6:31. doi: 10.1186/1741-7015-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]