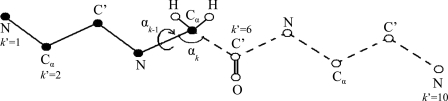
Figure 2.
An illustration of the HSMD reconstruction process of conformation i of a peptide consisting of three glycine residues, where for simplicity the oxygens and most of the hydrogens are discarded. At step k′ = 6, the TPs related to the “past” atoms k′ = 1, …, 5 (depicted by full spheres) have already been determined and these atoms are kept fixed in their positions at conformation i. At this step (6) one calculates the TPs of bond angle αk (defined by C′-Cα-N) and dihedral angle αk-1 (defined by C′-Cα-N-C′) which are related to C′, where k = 2k′ and thus k = 12. The TPs are obtained from an MD simulation where the as yet unreconstructed atoms k′ = 6, …, 10 (the “future” atoms) are moved (depicted by empty spheres connected by dashed lines) while the past atoms k′ = 1, …, 5 are kept fixed; note that the future part should remain within the limits of the microstate and future-past interactions are taken into account. Small bins δαk-1 and δαk are centered at the values αk-1 and αk in i. The TP is calculated from the number of visits [nvisit, Eq. 11] of the future part to δαk-1 and δαk simultaneously during the simulation. After ρHS (α11 α12|α10 ,...,α1) [Eq. 12] has been determined, the coordinates of C′ (and O) are fixed at their positions in i, i.e., they become “past” atoms and the process continues.