Abstract
Background
Reproductive character displacement (RCD) is a common and taxonomically widespread pattern. In marine broadcast spawning organisms, behavioral and mechanical isolation are absent and prezygotic barriers between species often operate only during the fertilization process. Such barriers are usually a consequence of differences in the way in which sperm and egg proteins interact, so RCD can be manifest as faster evolution of these proteins between species in sympatry than allopatry. Rapid evolution of these proteins often appears to be a consequence of positive (directional) selection. Here, we identify a set of candidate gamete recognition proteins (GRPs) in the ascidian Ciona intestinalis and showed that these GRPs evolve more rapidly than control proteins (those not involved in gamete recognition). Choosing a subset of these gamete recognition proteins that show evidence of positive selection (CIPRO37.40.1, CIPRO60.5.1, CIPRO100.7.1), we then directly test the RCD hypothesis by comparing divergence (omega) and polymorphism (McDonald-Kreitman, Tajima's D, Fu and Li's D and F, Fay and Wu's H) statistics in sympatric and allopatric populations of two distinct forms of C. intestinalis (Types A and B) between which there are strong post-zygotic barriers.
Results
Candidate gamete recognition proteins from two lineages of C. intestinalis (Type A and B) are evolving more rapidly than control proteins, consistent with patterns seen in insects and mammals. However, ω (dN/dS) is not significantly different between the sympatric and allopatric populations, and none of the polymorphism statistics show significant differences between sympatric and allopatric populations.
Conclusions
Enhanced prezygotic isolation in sympatry has become a well-known feature of gamete recognition proteins in marine broadcast spawners. But in most cases the evolutionary process or processes responsible for this pattern have not been identified. Although gamete recognition proteins in C. intestinalis do appear to evolve more rapidly, on average, than proteins with other functions, rates of evolution are not different in allopatric and sympatric populations of the two reproductively isolated forms. That sympatry is probably human-mediated, and therefore recent, may explain the absence of RCD.
Background
Reproductive isolation between incipient species is of particular relevance to the process of speciation. Reproductive character displacement - 'the pattern of greater divergence of a (prezygotic) isolating trait in areas of sympatry between closely related taxa than in areas of allopatry' [1,2] is a common and taxonomically widespread pattern which is of great interest when studying reproductive isolation [3]. Evidence for RCD comes from groups as diverse as fungi [4,5], plants [6], insects [7,8] mollusks [9], fish [10,11] and amphibians [12]. However, the majority of RCD examples come from the Drosophila literature [13-17].
The study of RCD has historically been tied to the process of reinforcement, the evolution of prezygotic isolation resulting from selection against hybrid individuals [13-15]. More recently, however, workers have emphasized that RCD can be caused by other factors, including ecological variables [18,19]. But even where selection has been shown to play a role in RCD [20,21], the specific action of this selection remains unknown.
RCD has been documented in several taxa of marine broadcast spawning organisms [18]. The study of RCD is often more tractable in broadcast spawners than in internal fertilizers. Broadcast spawning individuals simultaneously release sperm and eggs into the water column, where fertilization occurs. Therefore, behavioral and mechanical isolation are absent in broadcast spawning organisms and if two sympatric broadcast spawning species are not temporally isolated [22], prezygotic barriers between these two species occur only during the fertilization process. Such barriers are usually a consequence of differences in the way in which sperm and egg proteins interact, so RCD is often manifest as faster evolution of these proteins between species in sympatry than allopatry [23,24]. Rapid evolution of these proteins appears to proceed through the action of positive (directional) selection, increasing the proportion of nonsynonymous substitutions (those that cause amino acid changes) relative to synonymous substitutions (the dn/ds ratio) [25].
New information has allowed us to use the broadcast spawning ascidian Ciona intestinalis, a longtime model in developmental biology, to study the early stages of RCD. C. intestinalis comprises two distinct and divergent lineages, now termed Type A and Type B [26-28], Type A is thought to be native to the Northwestern Pacific Ocean and to have invaded the Eastern Pacific Ocean, the Mediterranean Sea, the Atlantic coast of South Africa, and the Black Sea [29,30]. Type B, native to the Northern Atlantic Ocean [31,32], has invaded the Western Atlantic Ocean [27]. The ranges of Type A and B overlap in the English Channel and the Atlantic coast of France, where limited introgression occurs between Type A and B individuals from these sympatric populations [26]. Hybrids from crosses between allopatric individuals are sterile or inviable in the laboratory [28], but the evidence for introgression in nature shows that these two lineages are not completely reproductively isolated. Nothing is yet known about pre or postzygotic barriers in sympatry.
Here, we first identify a set of candidate gamete recognition proteins (GRPs) from C. intestinalis using proteomic and bioinformatic techniques and test whether these proteins evolve more rapidly than control proteins. Then, choosing a subset of these proteins that show evidence of positive selection, we directly test the RCD hypothesis by comparing divergence and polymorphism statistics in sympatric and allopatric populations of Type A and B C. intestinalis. We ask whether signatures of positive selection are stronger in sympatric than allopatric populations.
Results
Identification of candidate GRPs from sperm: proteomics
161 sperm proteins were identified by LC-MS/MS (Liquid Chromatography-Mass Spectrometry/Mass Spectrometry). Each of these proteins was subsequently identified using one or more of the following sections of the CIPRO database (Ciona intestinalis Protein): the descriptive summary available for many proteins, the Pfam Domain search, and the BlastP search, or the GO (Gene Ontology) program. 144 of the proteins are unlikely to be GRPs; they are likely involved in the movement or metabolism of the sperm (e.g. actin, dynein, myosin, tektin, α-tubulin, ATP-synthase, creatine kinase, enolase, malate dehydrogenase). The identities of these 144 proteins are available from the authors. Of the remaining 17 proteins, seven were likely GRPs, and 10 could not be identified as similar to any known proteins. Of these 17 proteins, four could not be analyzed because primers based on the Type A genome sequence would not amplify the homolog from Type B individuals. An additional gene could not be analyzed because the gene encoding the protein did not have significant tblastn hits to the Type A genome (and therefore no primers could be designed). We selected the remaining 12 proteins for further analysis, 3 of which were likely GRPs.
Identification of candidate GRPs proteins from sperm: bioinformatics
We also used a bioinformatic approach to identify potential GRPs. First, we accessed the functional classifications of Type A testis ESTs sequenced by [33] and selected 25 ESTs that might code for GRPs. We then located the genes corresponding to these ESTs with the KOG (EuKaryotic Orthologous Groups) tool provided on JGI's (Joint Genome Institute) C. intestinalis genome browser http://genome.jgi-psf.org/Cioin2/Cioin2.home.html. These genes were then searched against the CIPRO database using blastx to identify resulting protein matches. 19 of these proteins were determined to be GRP candidates, but 10 failed to amplify and/or sequence in Type B individuals; the remaining nine were selected for further study.
Second, we located every protein in the CIPRO database that was identified as being expressed only in testes. We chose a subset of 10 of these proteins that were likely GRPs, of which nine could be amplified from Type B cDNA. In total, we selected 12 candidate GRPs identified proteomically and 18 identified bioinformatically. We also selected 9 control proteins (not involved in the fertilization process) from the proteomic analyses to compare with the putative GRPs. These control proteins, CIPRO129.24.1, CIPRO183.42.1, CIPRO2.134.1, CIPRO32.59.1, CIPRO53.35.1, CIPRO57.34.1, CIPRO68.37.1, CIPRO81.38.1 and CIPRO963.2.1, were selected because Pfam Domain and BlastP Searches in CIPRO identified them as proteins involved in basic cellular processes rather than fertilization (e.g. tubulin, NADH dehydrogenase, aconitate hydratase).
Comparison of dN/dS values between candidate GRPs and control proteins
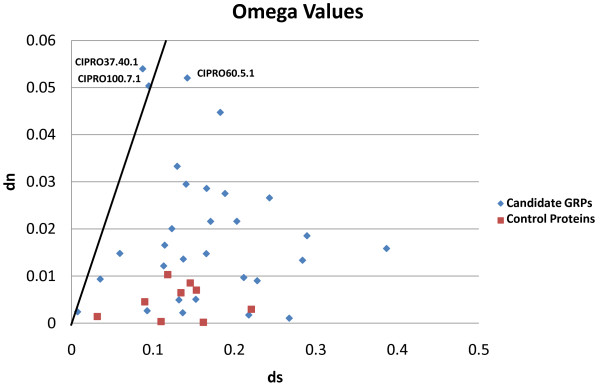
The dN/dS values for candidate GRPs are significantly higher than the dN/dS values for control proteins (Figure 1, p = 0.004891 using a one-tailed Mann-Whitney U Test).
Figure 1.
Pairwise omega (dN/dS) values. This graph shows pairwise dN/dS values (Type A vs. Type B C. intestinalis) for 30 candidate gamete recognition proteins (GRPs) and 9 control proteins. The line is dN/dS = 0.5.
However, PAML analyses assume that dS values are constant across a sequence. If some sites across the sequence have unusually low dS values, a high dN/dS value could be inferred in the absence of positive selection [34]. Thus, significantly higher dN/dS values for candidate gamete recognition genes than for control genes could be the result of either higher dN or lower dS values in the candidate recognition genes, and only higher dN values provide evidence of positive selection.
To address this issue, we performed two-tailed Mann-Whitney U test in R (the Shapiro-Wilk test found evidence for non-normality), comparing dN values in candidate gamete recognition vs. control genes, and dS values in candidate gamete recognition vs. control genes. dN values were significantly different between candidate gamete recognition and control genes (p = 0.002), whereas dS values were not (p = 0.269). These tests are consistent with the assumption that dN, rather than dS, is driving the observed pattern.
Figure 1 also shows that two proteins have a dN/dS ratio greater than 0.5, which is the value above which we consider positive selection likely to be occurring when conservative pairwise dN/dS comparisons are used. A study by [35] across many different taxa showed that if a pairwise comparison yielded a dN/dS ratio of greater than 0.5, evidence for selection was subsequently observed when more sensitive site-specific tests were used. The rationale here is that dN/dS < 1 can still indicate positive selection at some subset of the amino acid residues within a protein, although much of the protein may be subject to constraint. Of the two proteins with dN/dS > 0.5, CIPRO37.40.1 was identified from the CIPRO database as having testis-only expression, and contains domains similar to ricin-type beta-trefoil lectin domains (dN/dS = 0.618, dN = 0.054, dS = 0.087).
CIPRO100.7.1 was identified proteomically, and its function is unknown. CIPRO100.7.1 is a large protein (1,225 amino acids), and was therefore divided into three sections for sequencing. Sections 2, 3, and all 3 sections analyzed together had dN/dS values less than 0.5, so only Section 1 was analyzed in subsequent sympatric vs. allopatric comparisons. Section 1 of CIPRO100.7.1 had a dN/dS equal to 0.531, a dN equal to 0.050, and a dS equal to 0.095. One protein, CIPRO60.5.1, has a dN/dS ratio lower than 0.5, but was identified in the CIPRO database as a metalloproteinase and has a GO biological function of "sperm binding to zona pellucida" (dN/dS = 0.366, dN = 0.052, dS = 0.142). We chose these three proteins for the sympatric vs. allopatric comparisons, either because they showed evidence of positive selection (in the case of CIPRO37.40.1 and CIPRO100.7.1), or because their putative function was so clearly related to gamete recognition (in the case of CIPRO60.5.1).
Sympatric vs. allopatric divergence analyses
The results of the divergence analyses are shown in Table 1. For all of the candidate GRPs and control proteins, the 95% credible interval of the distribution of the differences between sympatric and allopatric ω (dN/dS) values included zero. This indicates that omega values were not significantly different between sympatry and allopatry for any of the proteins examined. Not enough variation was present in the sympatric Type A alleles for CIPRO60.5.1 or mtCOI to obtain reliable omega values, so the sympatric Type A vs. allopatric Type A comparison could not be performed for these proteins.
Table 1.
Sympatric vs. allopatric comparisons of omega values
| Candidate GRPs | Posterior mean of the difference between estimated omega values | 95% credible interval of the difference between estimated omega values |
|---|---|---|
| CIPRO37.40.1 Sympatric A vs. Allopatric A | -0.52 | -1.865 to 0.779 |
| CIPRO37.40.1 Sympatric B vs. Allopatric B | 0.01 | -0.572 to 0.463 |
| CIPRO60.5.1 Sympatric A vs. Allopatric A | NA (Not enough variation in sympatric Type A) | NA (Not enough variation in sympatric Type A) |
| CIPRO60.5.1 Sympatric B vs. Allopatric B | -0.09 | -0.48 to 0.35 |
| CIPRO100.7.1 Sympatric A vs. Allopatric A | 0.29 | -0.471 to 0.939 |
| CIPRO100.7.1 Sympatric B vs. Allopatric B | 0.01 | -0.771 to 0.651 |
| Control Proteins | Posterior mean of the difference between estimated omega values | 95% credible interval of the difference between estimated omega values |
| CIPRO53.35.1 Sympatric A vs. Allopatric A | 0 | -0.015 to 0.0002 |
| CIPRO53.35.1 Sympatric B vs. Allopatric B | 0.19 | -0.017 to 0.443 |
| mtCOI Sympatric A vs. Allopatric A | NA (Not enough variation in sympatric Type A) | NA (Not enough variation in sympatric Type A) |
| mtCOI Sympatric B vs. Allopatric B | 0.26 | -0.064 to 0.791 |
Sympatric vs. allopatric polymorphism analyses
The summary statistics are shown in Table 2. No consistent differences between sympatric and allopatric Type A or between sympatric and allopatric Type B can be discerned for any of these statistics for any of these candidate genes. Table 3 presents the results of the McDonald-Kreitman tests. Fixed nonsynonymous substitutions are not more common in sympatric than allopatric comparisons; these tests provide no evidence for positive selection on the genes encoding these three candidate GRPs in sympatry. None of the statistics for which significance was determined by coalescent simulation (D, D*, F*, H) showed significant differences between sympatry and allopatry for any of the genes examined, as the confidence intervals for sympatric populations always contained the mean values for allopatric populations, and vice versa (Table 4 for Tajima's D, Table 5 for D* and F*, Table 6 for H).
Table 2.
Summary Statistics.
| Candidate GRP genes | Population | n | θ | π (total) | π (synonymous sites) | π (nonsynonymous sites) |
|---|---|---|---|---|---|---|
| CIPRO37.40.1 | Sympatric Type A | 28 | 0.026 | 0.022 | 0.042 | 0.016 |
| Allopatric Type A | 24 | 0.013 | 0.012 | 0.013 | 0.010 | |
| Sympatric Type B | 16 | 0.019 | 0.016 | 0.041 | 0.008 | |
| Allopatric Type B | 8 | 0.043 | 0.050 | 0.109 | 0.032 | |
| CIPRO60.5.1 | Sympatric Type A | 20 | 0.002 | 0.002 | 0.000 | 0.003 |
| Allopatric Type A | 22 | 0.024 | 0.018 | 0.030 | 0.015 | |
| Sympatric Type B | 24 | 0.035 | 0.034 | 0.057 | 0.025 | |
| Allopatric Type B | 20 | 0.021 | 0.026 | 0.042 | 0.019 | |
| CIPRO100.7.1 | Sympatric Type A | 24 | 0.011 | 0.009 | 0.017 | 0.005 |
| Allopatric Type A | 26 | 0.007 | 0.007 | 0.012 | 0.004 | |
| Sympatric Type B | 22 | 0.018 | 0.020 | 0.031 | 0.015 | |
| Allopatric Type B | 8 | 0.017 | 0.022 | 0.030 | 0.017 | |
| Control genes | ||||||
| CIPRO53.35.1 | Sympatric Type A | 22 | 0.008 | 0.006 | 0.027 | 0.001 |
| Allopatric Type A | 32 | 0.006 | 0.005 | 0.020 | 0.000 | |
| Sympatric Type B | 32 | 0.013 | 0.011 | 0.043 | 0.002 | |
| Allopatric Type B | 28 | 0.016 | 0.015 | 0.042 | 0.007 | |
| mtCOI | Sympatric Type A | 10 | 0.000 | 0.000 | 0.000 | 0.000 |
| Allopatric Type A | 12 | 0.006 | 0.004 | 0.021 | 0.000 | |
| Sympatric Type B | 9 | 0.005 | 0.005 | 0.023 | 0.000 | |
| Allopatric Type B | 11 | 0.005 | 0.005 | 0.021 | 0.001 | |
n is the number of alleles sequenced from each population.
Table 3.
Results of the McDonald Kreitman Tests for all genes.
| Candidate GRP genes | Test | FS | PS | FN | PN | P value (Fisher's Exact Two Tailed Test) |
|---|---|---|---|---|---|---|
| CIPRO37.40.1 | Sympatric Type A vs. Sympatric Type B | 0 | 35 | 0 | 36 | NA |
| Allopatric Type A vs. Allopatric Type B | 9 | 38 | 15 | 48 | 0.64 | |
| CIPRO60.5.1 | Sympatric Type A vs. Sympatric Type B | 6 | 27 | 7 | 22 | 0.76 |
| Allopatric Type A vs. Allopatric Type B | 3 | 16 | 6 | 19 | 0.71 | |
| CIPRO100.7.1 | Sympatric Type A vs. Sympatric Type B | 9 | 25 | 12 | 30 | 1 |
| Allopatric Type A vs. Allopatric Type B | 5 | 12 | 8 | 17 | 1 | |
| Control genes | Test | FS | PS | FN | PN | P value (Fisher's Exact Two Tailed Test) |
| CIPRO53.35.1 | Sympatric Type A vs. Sympatric Type B | 0 | 18 | 0 | 7 | NA |
| Allopatric Type A vs. Allopatric Type B | 4 | 17 | 1 | 11 | 0.63 | |
| mtCOI | Sympatric Type A vs. Sympatric Type B | 76 | 9 | 3 | 1 | 0.38 |
| Allopatric Type A vs. Allopatric Type B | 70 | 21 | 3 | 2 | 0.59 | |
FS = Fixed Synonymous. PS = Polymorphic Synonymous. FN = Fixed Nonsynonymous. PN = Polymorphic Nonsynonymous
Table 4.
Tajima's D statistics for all genes
| Candidate GRP genes | Population | D statistic | P-value | 95% confidence interval |
|---|---|---|---|---|
| CIPRO37.40.1 | Sympatric Type A | -0.078 | 0.281 | "-1.68841 to 1.69923" |
| Allopatric Type A | -0.004 | 0.157 | "-1.20834 to 1.17" | |
| Sympatric Type B | -0.132 | 0.290 | "-1.77 to 1.611" | |
| Allopatric Type B | 0.006 | 0.092 | "-0.99686 to 0.94395" | |
| CIPRO60.5.1 | Sympatric Type A | -0.018 | 0.541 | "-1.51 to 1.66" |
| Allopatric Type A | -0.087 | 0.561 | "-1.69367 to 1.76737" | |
| Sympatric Type B | -0.058 | 0.528 | "-1.45443 to 1.32636" | |
| Allopatric Type B | 0.006 | 0.519 | "-1.54796 to 1.72153" | |
| CIPRO100.7.1 | Sympatric Type A | -0.018 | 0.503 | "-1.46162 to 1.35414" |
| Allopatric Type A | 0.009 | 0.510 | "-1.3292 to 1.48969" | |
| Sympatric Type B | -0.014 | 0.515 | "-0.92013 to 0.96708" | |
| Allopatric Type B | -0.042 | 0.526 | "-1.35751 to 1.17643" | |
| Control genes | ||||
| CIPRO53.35.1 | Sympatric Type A | -0.057 | 0.544 | "-1.68808 to 1.83296" |
| Allopatric Type A | -0.002 | 0.511 | "-1.37399 to 1.4227" | |
| Sympatric Type B | -0.027 | 0.524 | "-1.42163 to 1.46487" | |
| Allopatric Type B | -0.061 | 0.545 | "-1.31043 to 1.28035" | |
| mtCOI | Sympatric Type A | -0.002 | 0.127 | "-1.11173 to 1.43863" |
| Allopatric Type A | -0.065 | 0.185 | "-1.83094 to 1.77946" | |
| Sympatric Type B | -0.064 | 0.203 | "-1.67754 to 1.75974" | |
| Allopatric Type B | -0.063 | 0.186 | "-1.75914 to 1.8452" | |
Table 5.
Fu and Li's D* and F* statistics for all genes
| Candidate GRP genes | Population | D* | P-value | 95% confidence interval | F* | P-value | 95% confidence interval |
|---|---|---|---|---|---|---|---|
| CIPRO37.40.1 | Sympatric Type A | -0.157 | 0.517 | "-2.59 to 1.25985" | -0.111 | 0.496 | "-2.498 to 1.52763" |
| Allopatric Type A | -0.028 | 0.670 | "-1.52677 to 1.239" | -0.063 | 0.589 | "-1.58919 to 1.21221" | |
| Sympatric Type B | -0.034 | 0.578 | "-2.24738 to 1.28647" | -0.090 | 0.540 | "-2.66286 to 1.49659" | |
| Allopatric Type B | -0.024 | 0.734 | "-1.00 to 0.87955" | 0.007 | 0.612 | "-1.03183 to 1.04566" | |
| CIPRO60.5.1 | Sympatric Type A | 0.032 | 0.458 | "-1.96617 to 1.25359" | -0.028 | 0.492 | "-2.16055 to 1.477" |
| Allopatric Type A | -0.040 | 0.432 | "-2.464 to 1.25307" | -0.086 | 0.469 | "-2.56615 to 1.49075" | |
| Sympatric Type B | -0.026 | 0.452 | "-1.83617 to 1.17832" | -0.076 | 0.491 | "-2.08 to 1.29" | |
| Allopatric Type B | -0.087 | 0.466 | "-2.08275 to 1.17893" | -0.071 | 0.471 | "-2.18321 to 1.38673" | |
| CIPRO100.7.1 | Sympatric Type A | -0.018 | 0.467 | "-1.83829 to 1.22556" | -0.024 | 0.472 | "-1.86692 to 1.35458" |
| Allopatric Type A | -0.056 | 0.497 | "-1.87585 to 1.23" | -0.070 | 0.506 | "-2.09829 to 1.46478" | |
| Sympatric Type B | 0.020 | 0.471 | "-1.1613 to 0.99970" | -0.037 | 0.512 | "-1.23262 to 1.02296" | |
| Allopatric Type B | -0.014 | 0.481 | "-1.33924 to 1.16179" | -0.076 | 0.542 | "-1.57039 to 1.29433" | |
| Control genes | |||||||
| CIPRO53.35.1 | Sympatric Type A | -0.024 | 0.472 | "-1.86692 to 1.35458" | 0.013 | 0.456 | "-2.20751 to 1.64394" |
| Allopatric Type A | -0.070 | 0.506 | "-2.09829 to 1.46478" | -0.012 | 0.501 | "-1.68033 to 1.45582" | |
| Sympatric Type B | -0.037 | 0.512 | "-1.23262 to 1.02296" | -0.057 | 0.504 | "-1.91043 to 1.46351" | |
| Allopatric Type B | -0.076 | 0.542 | "-1.57039 to 1.29433" | -0.004 | 0.474 | "-1.88842 to 1.46422" | |
| mtCOI | Sympatric Type A | 0.008 | 0.762 | "-1.24341 to 1.02623" | -0.039 | 0.786 | "-1.34668 to 1.06879" |
| Allopatric Type A | -0.006 | 0.440 | "-2.229 to 1.40344" | -0.102 | 0.487 | "-2.39875 to 1.54664" | |
| Sympatric Type B | -0.048 | 0.463 | "-1.92508 to 1.43324" | -0.092 | 0.486 | "-2.10684 to 1.58013" | |
| Allopatric Type B | -0.093 | 0.489 | "-2.07471 to 1.42077" | -0.065 | 0.469 | "-2.30147 to 1.50465" | |
Table 6.
Fay and Wu's H test for all genes.
| Candidate GRP genes | Population | H statistic | P-value | 95% confidence interval |
|---|---|---|---|---|
| CIPRO37.40.1 | Sympatric Type A | -0.166 | 0.348 | "-21.476 to 7.34392" |
| Allopatric Type A | -0.040 | 0.434 | "-6.39 to 4.01449" | |
| Sympatric Type B | 0.114 | 0.333 | "-16.6833 to 6.3" | |
| Allopatric Type B | 0.233 | 0.430 | "-16.64 to 11.64" | |
| CIPRO60.5.1 | Sympatric Type A | 0.006 | 0.351 | "-2.37427 to 1.222" |
| Allopatric Type A | -0.216 | 0.338 | "-20.5 to 7.41126" | |
| Sympatric Type B | -0.144 | 0.394 | "-18.956 to 9.159" | |
| Allopatric Type B | -0.161 | 0.392 | "-13.53684 to 5.69474" | |
| CIPRO100.7.1 | Sympatric Type A | 0.080 | 0.387 | "-6.28261 to 3.78261" |
| Allopatric Type A | 0.014 | 0.401 | "-4.61538 to 2.75692" | |
| Sympatric Type B | 0.174 | 0.436 | "-8.8658 to 6.02597" | |
| Allopatric Type B | -0.055 | 0.425 | "-11.00 to 6.07143" | |
| Control genes | ||||
| CIPRO53.35.1 | Sympatric Type A | -0.061 | 0.347 | "-5.36797 to 2.07792" |
| Allopatric Type A | 0.003 | 0.413 | "-2.61694 to 1.54839" | |
| Sympatric Type B | 0.017 | 0.420 | "-4.03226 to 2.54839" | |
| Allopatric Type B | -0.027 | 0.406 | "-6.68783 to 3.35979" | |
| mtCOI | Sympatric Type A | -0.001 | 0.718 | "-1.06667 to 0.4444" |
| Allopatric Type A | 0.024 | 0.312 | "-6.72727 to 2.4242" | |
| Sympatric Type B | -0.044 | 0.344 | "-6.47222 to 2.58333" | |
| Allopatric Type B | -0.119 | 0.315 | "-8.69091 to 2.7272" | |
Discussion
Comparison of dN/dS values between candidate GRPs and control proteins
Candidate GRPs in C. intestinalis are evolving more rapidly than control proteins, and this pattern is likely driven by substitutions at nonsynonymous sites. Rapid evolution has been documented at specific GRPs (i.e. lysin, VERL and bindin) in marine broadcast spawners, and dN/dS values are lower for mtCOI than the GRPs lysin and VERL for green and pink abalone [36]. The pattern we see in C. intestinalis has also been documented in insects and mammals (e.g. butterflies: [37], field crickets: [38], mouse and human: [39], primates: [40]). This study suggests that a pattern of faster evolution in reproductive proteins may apply to external as well as internal fertilizers.
One explanation for faster evolution in candidate GRPs than control proteins is sperm competition, which occurs in Ciona as it does in internal fertilizers. Selection could be acting on any proteins that determine how quickly sperm fertilize eggs: proteins involved in metabolism, motility, binding, penetration, etc. However, as Figure 1 shows that candidate GRPs are evolving more rapidly than sperm proteins that are not candidate GRPs (control proteins), proteins directly involved in sperm-egg interactions are more likely to be experiencing directional selection than those involved in facilitating sperm access to the egg.
Rapid evolution of sperm GRPs might also result from sexual conflict, which occurs when the optimal outcomes of fertilization are different for sperm and eggs. For sperm, the optimal outcome is fertilization of an egg as quickly as possible. But in many taxa, fertilization of eggs by multiple sperm (polyspermy) results in developmental defects. Therefore, the optimal outcome of fertilization for an egg may be slower fertilization, to avoid polyspermy [25]. Ascidians like C. intestinalis often live in close proximity to many conspecific individuals [41], and an individual usually sends sperm into the water column before eggs (ascidians are hermaphrodites). So eggs are usually released into a vast amount of sperm spawned from many neighbors, making the risk of polyspermy very high. Perhaps in response to this risk, ascidians have evolved two separate blocks to polyspermy, whereas many other marine broadcast spawners have a single block [41]. Given these effective polyspermy blocks, sexual conflict resulting from polyspermy is not likely to be major driver of GRP evolution in ascidians.
Reinforcement could be driving the enhanced prezygotic isolation in sympatry. We know that hybrids between allopatric populations Type A and B C. intestinalis are sterile or inviable in the laboratory, so selection could favor GRPs that allow sperm to preferentially bind to conspecific eggs. However, reinforcing selection could be counteracted by gene flow from allopatric populations [42], especially when secondary contact is recent. Pairwise FST calculations between sympatric and allopatric Type A, and sympatric and allopatric Type B populations show that these populations are not significantly differentiated at any of the three genes encoding the GRPs (data not shown); migration may therefore be occurring between sympatric and allopatric populations.
Lastly, egg surface proteins could be changing rapidly to prevent pathogens from entering the egg. If the same proteins involved in preventing microbial attack are involved in sperm/egg recognition, this could lead to the rapid evolution of sperm proteins to keep up with the ever-changing egg proteins.
Evolution of candidate GRPs in Ciona intestinalis - no evidence for RCD
Our data provide no evidence that positive selection is enhanced in sympatry, and if these candidate GRPs are involved in prezygotic isolation, we have no evidence for enhanced prezygotic isolation. The polymorphism statistics likewise give no indication that RCD is occurring in these three proteins.
We cannot necessarily conclude from lack of evidence for RCD in CIPRO37.40.1, CIPRO60.5.1 and CIPRO100.7.1 that RCD is not occurring in this system. If enhanced prezygotic isolation between Type A and B does exist, there are several reasons why we might not have detected it in this study. First, primers for candidate GRPs were developed from the Type A genomic sequence and were used to amplify and sequence both Type A and B individuals (the Type B genome has not been sequenced). But Type A and B are substantially divergent (p-distances: 0.124 at mtCOI, 0.035 to 0.116 for six nuclear loci; [43]), which could explain why 15 genes encoding GRP candidates could not be successfully amplified and/or sequenced in Type B individuals. It is possible that the genes that could not be amplified and/or sequenced (and were therefore excluded from the analyses) encode proteins that are evolving more rapidly between Type A and B than those that were included in the analyses. If this is the case, we may have missed proteins for which dN/dS values are >1, proteins that would have been included in the sympatric vs. allopatric tests of RCD.
It is, of course, also possible that the three proteins that have high dN/dS values are themselves not involved in gamete recognition. While some aspects of the fertilization process in solitary ascidians such as C. intestinalis are well-characterized [44,45], the genes and corresponding proteins responsible for species-specificity have not been identified, as they have in other marine broadcast spawners [23,24].
Two sperm proteins that interact directly with the egg have been identified in C. intestinalis, but it is not known whether these proteins are involved in gamete recognition. The first protein is α L-fucosidase, which binds to the vitelline coat of the egg [46]. Five of our candidate GRPs (CIPRO187.4.1, CIPRO19.75.1, CIPRO33.15.1, CIPRO552.7.1, and CIPRO58.12.1) had domains also found in α L-fucosidases, but none of these proteins were expressed in testis tissue, based on expression data in CIPRO. CIPRO187.4.1 could not be amplified from Type B, and the other four genes did not have dN/dS ratios > 0.5. A second protein involved in sperm-egg interactions is a chymotrypsin-like enzyme that may dissolve the vitelline coat of the egg [47]. However, the amino acid sequence for this protein is not available and we identified dozens of chymotrypsin-like proteins in the genome.
Temporal isolation of Types A and B could also contribute to prezygotic barriers. We do not know whether Type A and B release gametes at the same time of day or in the same season in the English Channel. Since some gene flow has occurred [26], spawning must at least partially overlap.
Gene flow between sympatric and allopatric populations could be obscuring RCD if it is occurring in this system. Pairwise FST calculations show that sympatric and allopatric populations of each type are not significantly differentiated, so gene flow between sympatric and allopatric populations is a possibility.
Finally, because secondary contact between Type A and B C. intestinalis in the English Channel may well be recent, RCD may not have yet taken place. Type B is native to Northern Europe and presumably a long-time resident of the English Channel. We do not know when Type A invaded the English Channel. The first published record of Type A in this area was in 2007 [26], but as Type A and B were only recognized in 2005 [48], Type A living in this area prior to 2005 would not have been distinguished from the native Type B. However, the introduction of Type A was likely human-mediated [26], and therefore a recent invasion on an evolutionary timescale. Evidence for RCD has been found in several systems where secondary contact is relatively recent. For example, RCD in mate choice is observed when limnetic and benthic sticklebacks co-occur in glacial lakes in British Columbia [20]. Similarly, RCD has been documented in the Mus musculus and Mus domesticus hybrid zone, which is thought to represent secondary contact during the Neolithic (since 9500 BCE) [49].
Positive selection on GRPs in other marine broadcast spawners: RCD
Some of the most rapidly evolving proteins yet discovered are GRPs in marine broadcast spawners (e.g. bindin in sea urchins, lysin in abalone and mussels). In sea urchins, the bindin protein facilitates sperm attachment to the egg and fusion of sperm and egg [50]. In three genera of sea urchin that include sympatric species (Echinometra, Heliocidaris, and Strongylocentrotus), regions of bindin show evidence of positive selection ([51] and references therein). In Arbacia, Lytechinus and Tripneustes, genera that do not contain sympatric species, bindin shows no evidence of positive selection ([51] and references therein). This pattern is consistent with RCD on bindin in sea urchins.
Stronger evidence for RCD comes from a study of Echinometra oblonga, which has populations that are sympatric and allopatric with Echinometra species C [23]. Substantial divergence in bindin alleles between E. oblonga and E. sp. C. occurs where the two species are sympatric, but not where they are allopatric [23].
In abalone and mussels, sperm proteins known as lysins are involved in dissolution of the egg vitelline envelope, enabling the sperm to enter the egg. The best-characterized lysins are in the abalone genus Haliotis. An early study of 19 sympatric Haliotis species (19 sympatric and 1 allopatric species) found many pairwise comparisons with dN/dS values > 1 [52]. A later study of 25 species corroborated the pairwise results of [52] and also used maximum likelihood models of codon substitution to identify lineage and site-specific evidence of positive selection [53]. Lineages containing sympatric or closely related species usually had dN/dS values > 1, whereas lineages with distantly related allopatric species always had dN/dS values < 1, a pattern consistent with RCD [53]. But the authors also note a dN/dS value > 1 for the two branches separating a group of Japanese species from two groups of Californian species; this speciation event was likely allopatric.
In the mussel Mytilus galloprovincialis, two divergent clades of Lysin-M7 have been found: G and GD [24]. Evidence of positive selection is seen between G and GD, and within GD [24]. The divergence between the two clades is the result of rapid evolution in the GD clade, and GD alleles are found at higher frequencies in sympatric populations of M. galloprovincialis (where it hybridizes with Mytilus edulis) than in allopatric populations [24].
Conclusions
Enhanced prezygotic isolation in sympatry has become a well-known feature of GRPs in marine broadcast spawners. But in most cases the evolutionary process or processes responsible for this pattern have not been identified. Differentiating between the processes that can lead to patterns of RCD will provide important insights into the process of speciation in marine broadcast spawners.
Methods
Identification of candidate GRPs from sperm: proteomics experiment
Sperm was collected from Type A individuals living in Santa Barbara, CA, filtered through a 70 μm nylon cell strainer (BD Biosciences) by centrifuging for 3 minutes at 3,000 rpm, and stored dry at -80°C. Sperm samples from several different individuals were later pooled and diluted 5-fold in phosphate buffer; the concentration of this dilution was determined to be 915 μg/ml. 500 μl of this diluted sperm was shipped to the University of Victoria Genome BC Proteomics Centre for the experiments described below.
9.5 M urea, 50 mM NH4HCO3 and 0.2% SDS were added to the sample, which was then sonicated. The proteins then underwent disulphide reduction and sulphydryl alkylation (200 mM DTT and 200 mM iodoacetamide) and were digested overnight at 37°C with 20 mg trypsin (Promega). Samples were subsequently cleaned with a cation exchange Cartridge Kit (Applied Biosystems).
Strong cation exchange chromatography
10 mM KH2PO4 (pH 2.7), 25% ACN buffer was added to the sample, which was then injected onto a Polysulphoethyl A strong cation exchange chromatography (SCX) column (PolyLC, Columbia, MD). The flow rate was set to 0.5 ml min-1. After equilibration, a 0-35% gradient of 10 mM KH2PO4, 25% ACN, 0.5 M KCl was added for 30 min. Each SCX fraction was then moved to autosampler vials (Dionex/LC Packings, Amsterdam).
One-dimensional reversed-phase chromatography with online mass spectrometry
A hybrid Quadruple-TOF LC-MS/MS mass spectrometer (QStar Pulsar I, MDS Sciex) with a nanoelectrospray ionization source (Proxeon, Odense, Denmark) was used to complete the liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analyses. A C18AQ Nano LC and a Zorbax C18 guard column (Agilent Technologies) performed the chromatographic separation. The ANALYST QS v. 1.1 software service pack (ABI MDS SCIEX, Concord, Canada) gathered the data.
Mass spectrometry data analyses
the information dependent acquisition file was viewed using ANALYST v. 1.1 software, and the peak lists were assembled with a built-in MASCOT script (1.6b16 ABI--Matrix Science Limited). Spectra with less than 10 peaks were discarded. MASCOT v. 2.0 (Matrix Science Limited) was used to analyze the data. Spectrometry data were searched against a database of amino acid sequences from the CIPRO (Ciona intestinalis Protein) database http://cipro.ibio.jp/2.5/.
Comparison of dN/dS values between candidate GRPs and control proteins
Of proteins identified by proteomic analysis, we selected 39 proteins (30 candidate GRPs and 9 control proteins) for further analysis using proteomic and bioinformatic approaches (see Results section). Genomic sequences for the genes encoding the 39 proteins were located by performing tblastn searches to the Ciona intestinalis Ensembl genome server http://uswest.ensembl.org/Ciona_intestinalis/Info/Index. Primers were developed in coding regions, and the partial or entire coding regions of all 39 proteins were sequenced from cDNA of two Type A and two Type B individuals, all from allopatric populations. Testis tissues from these four individuals were collected in 2008 and immediately placed in RNAlater (Ambion). The tissue/RNAlater was frozen at -80°C within seven days of collection.
Total RNA was extracted from testis tissue with the RNAdvance Kit (Agencourt) and was used to synthesize single-stranded cDNA using SuperScript III reverse transcriptase (Invitrogen) and an oligo (dT) primer. A 5-fold dilution of the single-stranded cDNA was then PCR-amplified with TRsa and TS-PCR primers. The resulting PCR product was diluted 50-fold and used as the template for amplification of the coding regions for the 30 candidate GRPs proteins and 9 control proteins. The amplified coding regions were incubated with 0.25 μl each of Exonuclease I and Shrimp Antarctic Phosphatase at 37°C for 30 min, followed by 90°C for 10 min. The products were purified using CleanSeq beads (Agencourt), and the purified products were sequenced with a Big Dye Terminator Cycle sequencing kit and an Automated 3730 DNA Analyzer (Applied Biosystems). Sequences were edited, trimmed and aligned with Aligner (CodonCode Corporation, Dedham, MA). Primers and cycling conditions are available from the authors.
Once the sequences were obtained, the codeml program in PAML 4.4 [54] was used to obtain pairwise dN/dS values for each Type A vs. Type B combination (Type A Individual #1 vs. Type B Individual #1, Type A Individual #1 vs. Type B Individual #2, Type A Individual #2 vs. Type B Individual #1, Type A Individual #2 vs. Type B Individual #2). The average dN/dS value for all four combinations was calculated for each gene, and the dN/dS values of the putative GRPs and control proteins were found to be distributed non-normally using the Shapiro-Wilk test in R (version 2.10.1). The Shapiro-Wilk test is the most robust test of non-normality for small to medium sample sizes [55,56]. The dN/dS values of the candidate GRPs and control proteins were therefore compared using a one-tailed Mann-Whitney U test in R (version 2.10.1).
Sympatric vs. allopatric divergence analyses
Samples were collected in 2005-2009 from allopatric and sympatric populations of Type A and Type B. The allopatric population of Type A was from Half Moon Bay, CA while sympatric populations of Type A were from Perros-Guirec, France and Concarneau, France. Allopatric populations of Type B were collected from Woods Hole, MA and Gosport, England.
To obtain genomic DNA, ovaries were dissected from freshly-collected individuals, cut into several pieces, immediately preserved in DMSO (dimethyl sulfoxide), and ultimately (within 10 d) stored at -80°C until needed. Total DNA was extracted from the ovaries using the Qiagen DNeasy® Tissue Kit (Qiagen Corporation, Santa Clarita, CA).
At least 10 individuals from each of 4 populations (allopatric Type A and B, sympatric Type A and B) were sequenced for the genes encoding three candidate GRPs: CIPRO37.40.1, CIPRO60.5.1, CIPRO100.7.1. The criteria for selecting these three candidate GRPs for the sympatric/allopatric comparison, from the 30 candidate GRPs used in the comparison of dN/dS values between GRPs and control proteins, are discussed in the Results section.
The same individuals were also sequenced for two control proteins: CIPRO53.35.1 and mitochondrial cytochrome oxidase I (mtCOI). Sequences from all five loci were deposited in GenBank (Accession Numbers HQ872081-HQ872454). A signature of enhanced selection in sympatry vs. allopatry could be due to selective processes or demographic processes (e.g. recent population growth). But demographic processes would affect all genes, not just candidate GRPs. So proteins not involved in the fertilization process (control proteins) were also subjected to divergence analyses. CIPRO53.35.1 was selected as a control protein because it contains a domain similar to a ribosomal L32 protein domain and is expressed in many different tissues; whereas mtCOI is an enzyme in the electron transport chain of the mitochondria. For all nuclear genes the PCR product for each locus was cloned using the pGEM®-T kit and up to eight clones were PCR-amplified and sequenced as described above to obtain both alleles. When only one allele was found in eight clones, the individual was assumed to be homozygous for that allele.
We used omegaMap version 0.5 [57] to determine whether omega values were significantly different for sympatric Type A vs. allopatric Type A populations and for allopatric Type A vs. allopatric Type B populations. We chose omegaMap over PAML for three reasons. First, omegaMap is designed to calculate omega values in the presence of recombination, and every one of the five genes (those encoding CIPRO37.40.1, CIPRO60.5.1, CIPRO100.7.1, CIPRO53.35.1, and mtCOI) has at least one population where intragenic recombination is present. Second, unlike PAML, omegaMap takes a population genetics approach, so we can use all the alleles we sequenced from each population in our calculation of omegaMap. Lastly, the Bayesian inference implemented in omegaMap provides a perfect framework for testing whether omega values are significantly different in sympatry vs. allopatry.
Using omegaMap, we calculated omega (dN/dS) values for sympatric Type A, sympatric Type B, allopatric Type A and allopatric Type B populations separately for each gene. We chose 250,000 iterations for each run, with thinning set to 1,000. We used an improper inverse distribution to specify priors for all parameters. Initial parameter values were μ = 0.1, κ = 3.0, ω = 0.5, ρ = 0.1. A constant model was used, so that all sites are assumed to have the same omega value. The number of iterations discarded as burnin varied across runs, but was determined by plotting the traces of μ and κ; iterations affected by the starting value of the parameter were discarded. Two independent runs were conducted for each population/gene (e.g. CIPRO37.40.1_Sympatric A Run 1 and 2). These two runs were combined in all cases, after it was determined that the mean and 95% credible interval for each parameter in the two runs matched closely.
After the omega values for each population/gene were estimated, we permuted the posterior distribution of CIPRO37.40.1 sympatric Type A omega values and the posterior distribution of CIPRO37.40.1 allopatric Type A omega values (for example), obtained a distribution of the differences between these values, and calculated the mean and 95% credible interval for this distribution. If the 95% credible interval contained zero, the estimated omega values were not significantly different. All calculations were done in R (version 2.10.1).
Sympatric vs. allopatric polymorphism analyses
Both alleles from at least 10 individuals from each of four populations (allopatric Type A and B, sympatric Type A and B) were sequenced for three candidate gamete recognition genes and two control genes as described in the "Sympatric vs. allopatric divergence analyses" section.
For each population and each gene, the summary statistics θ and π were calculated in DnaSP 5.10.1 [58]. We also employed the following tests: McDonald-Kreitman [59], Tajima's D [60], Fu and Li's D* and F* [61], and Fay and Wu's H [62] in DnaSP. Statistical significance of D, D*, F* and H were determined using 1,000 coalescent simulations in DnaSP. Estimates of per gene recombination for each population were made in DnaSP and were then imported into the simulations. 95% confidence intervals for D, D*, F* and H statistics were also recorded; sympatric and allopatric populations were determined to be significantly different for each statistic if the confidence intervals of the sympatric population did not contain the mean of the allopatric population.
Authors' contributions
MLN collected the samples, generated the data, performed the analyses and wrote the manuscript. RGH provided funding, and assisted with the design of the study and the editing of the manuscript. Both authors have read and approved the final manuscript.
Contributor Information
Marie L Nydam, Email: mln32@cornell.edu.
Richard G Harrison, Email: rgh4@cornell.edu.
Acknowledgements
We would like to thank Erin Newman-Smith at the Ascidian Stock Center (supported by grant # R24GM075049 from the National Institutes of Health) for supplying animals, the Smith Laboratory at UCSB and John Bishop at Plymouth MBA for providing logistical assistance, Dr. Kazuo Inaba for sharing information about the CIPRO database, Derek Smith and the staff of the University of Victoria Genome BC Proteomics Centre for generating the proteomics data, members of the Harrison Laboratory and Jose Andres for advice, and two anonymous reviewers for critiquing the manuscript. omegaMap was accessed through the Computational Biology Service Unit's Web Computing Resources at Cornell University. This work was funded by NSF Doctoral Dissertation Grant DEB-0907862 to M.L.N. and R.G.H. and NSF Grant DEB-0639904 to R.G.H.
References
- Brown WL, Wilson EO. Character displacement. Syst Zool. 1956;5:49–64. doi: 10.2307/2411924. [DOI] [Google Scholar]
- Howard DJ. In: Hybrid Zones and the Evolutionary Process. Harrison RG, editor. Oxford University Press, New York; 1993. Reinforcement: the origin, dynamics, and fate of an evolutionary hypothesis; pp. 46–69. [Google Scholar]
- Rice WR, Hostert EE. Laboratory experiments on speciation: What have we learned in 40 years? Evolution. 1993;47:1637–1653. doi: 10.2307/2410209. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. Reproductive isolation and phylogenetic divergence in Neurospora: Comparing methods of species recognition in a model eukaryote. Evolution. 2003;57:2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. [DOI] [PubMed] [Google Scholar]
- Le Gac M, Giraud T. Existence of a pattern of reproductive character displacement in Homobasidiomycota but not in Ascomycota. J Evol Biol. 2008;21:761–772. doi: 10.1111/j.1420-9101.2008.01511.x. [DOI] [PubMed] [Google Scholar]
- Kay K, Schemske DW. Natural selection reinforces speciation in a radiation of neotropical forest plants. Evolution. 2008;62:2628–2642. doi: 10.1111/j.1558-5646.2008.00463.x. [DOI] [PubMed] [Google Scholar]
- Cooley JR, Marshall DC, Hill KBR, Simon C. Reconstructing asymmetrical reproductive character displacement in a periodical cicada contact zone. J Evol Biol. 2006;19:855–868. doi: 10.1111/j.1420-9101.2005.01056.x. [DOI] [PubMed] [Google Scholar]
- Nosil PB, Crespi BJ, Sandoval CP. Reproductive isolation driven by the combined effects of ecological adaptation and reinforcement. Proc R Soc Lond B. 2003;270:1911–1918. doi: 10.1098/rspb.2003.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda Y, Kawakita A, Kato M. Reproductive character displacement in genital morphology in Satsuma land snails. Am Nat. 2009;173:689–697. doi: 10.1086/597607. [DOI] [PubMed] [Google Scholar]
- Albert AYK, Schluter D. Reproductive character displacement of male stickleback mate preference: reinforcement or direct selection? Evolution. 2004;58:1099–1107. doi: 10.1111/j.0014-3820.2004.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Gabor CR, Ryan MJ. Geographical variation in reproductive character displacement in mate choice by sailfin mollies. Proc R Soc Lond B. 2001;268:1063–1070. doi: 10.1098/rspb.2001.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon EM. Diversification of conspecific signals in sympatry: geographic overlap drives multidimensional reproductive character displacement in frogs. Evolution. 2009;63:1155–1170. doi: 10.1111/j.1558-5646.2009.00650.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. "Patterns of speciation in Drosophila revisited". Evolution. 1997;51:295–303. doi: 10.2307/2410984. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.2307/2409213. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. New York: Columbia University Press; 1937. [Google Scholar]
- Higgie M, Blows M. The evolution of reproductive character displacement conflicts with how sexual selection operates within a species. Evolution. 2008;62:1192–1203. doi: 10.1111/j.1558-5646.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- Noor MAF. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland: Sinauer; 2004. [Google Scholar]
- Noor MAF. Reinforcement and other consequences of sympatry. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Schluter D. Reinforcement of stickleback mating preferences: Sympatry breeds contempt. Evolution. 1998;52:200–208. doi: 10.2307/2410935. [DOI] [PubMed] [Google Scholar]
- Saetre GP, Moum T, Bures S, Kral M, Adamjan M, Moreno J. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature. 1997;387:589–592. doi: 10.1038/42451. [DOI] [Google Scholar]
- Levitan DR, Fukami H, Jara J, Kline D, McGovern TM, McGhee KE, Swanson CA, Knowlton N. Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution. 2004;58:308–323. [PubMed] [Google Scholar]
- Geyer LB, Palumbi SR. Reproductive character displacement and the genetics of gamete recognition in tropical sea urchins. Evolution. 2003;57:1049–1060. doi: 10.1111/j.0014-3820.2003.tb00315.x. [DOI] [PubMed] [Google Scholar]
- Springer SA, Crespi BJ. Adaptive gamete-recognition divergence in a hybridizing Mytilus population. Evolution. 2007;61:772–783. doi: 10.1111/j.1558-5646.2007.00073.x. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Reproductive protein evolution. Ann Rev Ecol Syst. 2002;33:161–179. doi: 10.1146/annurev.ecolsys.33.010802.150439. [DOI] [Google Scholar]
- Nydam ML, Harrison RG. Introgression despite substantial divergence in a broadcast spawning marine invertebrate. Evolution. 2010. in press . [DOI] [PubMed]
- Nydam ML, Harrison RG. Genealogical relationships within and among shallow-water Ciona species (Ascidiacea) Mar Biol. 2007;151:1839–1847. doi: 10.1007/s00227-007-0617-0. [DOI] [Google Scholar]
- Caputi L, Andreakis N, Mastrototaro F, Cirino P, Vassillo M, Sordino P. Cryptic speciation in a model invertebrate chordate. Proc Natl Acad Sci USA. 2007;104:9364–9369. doi: 10.1073/pnas.0610158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Name W. The north and south American ascidians. B Am Mus Nat Hist. 1945;84:1–476. [Google Scholar]
- Kott P. The ascidians of Australia. Aus J Mar Fresh Res. 1952;3:206–233. [Google Scholar]
- Linne C. Vindobonae: Typis Ioannis Thomae, 1767-1770. Systema Naturae. 1767.
- Monniot C, Monniot F. Additions to the inventory of eastern tropical Atlantic ascidians; arrival of cosmopolitan species. Bull Mar Sci. 1994;54:71–93. [Google Scholar]
- Inaba K, Padma P, Satouh Y, Shin-I T, Kohara Y, Satoh N, Satou Y. EST analysis of gene expression in testis of the ascidian Ciona intestinalis. Mol Repro Dev. 2002;62:431–445. doi: 10.1002/mrd.10131. [DOI] [PubMed] [Google Scholar]
- Pond SK, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subject to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLOS Genetics. 2009. p. e:1000570. [DOI] [PMC free article] [PubMed]
- Walters JR, Harrison RG. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Mol Biol Evol. 2010. [DOI] [PubMed]
- Andres JA, Maroja LS, Harrison RG. Searching for candidate speciation genes using a proteomic approach: seminal proteins in field crickets. Proc R Soc Lond B. 2008;275:1975–1983. doi: 10.1098/rspb.2008.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Kulathinal RJ, Singh RS. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol Biol Evol. 2002;19:1973–1980. doi: 10.1093/oxfordjournals.molbev.a004021. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Lambert CC, Goudeau H, Franchet C, Lambert G, Goudeau M. Ascidian eggs block polyspermy by two independent mechanisms: one at the plasma membrane, the other involving the follicle cells. Mol Reprod Dev. 1997;48:137–143. doi: 10.1002/(SICI)1098-2795(199709)48:1<137::AID-MRD16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bigelow RS. Hybrid zones and reproductive isolation. Evolution. 1965;19:449–458. doi: 10.2307/2406242. [DOI] [Google Scholar]
- Nydam ML, Harrison RG. Polymorphism and divergence within the ascidian genus Ciona. Mol Phyl Evol. 2010;56:718–726. doi: 10.1016/j.ympev.2010.03.042. [DOI] [PubMed] [Google Scholar]
- Satoh N. Developmental Biology of Ascidians. 1. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- Sawada H, Yokosawa H, Lambert CC, eds. The Biology of Ascidians. 1. New York: Springer; 2000. [Google Scholar]
- Hoshi M, De Santis R, Pinto R, Cotelli F, Rosati F. Sperm glycosidases as mediators of sperm-egg interaction in the ascidians. Zool Sci. 1985;2:65–69. [Google Scholar]
- Marino R, De Santis R, Hirohashi N, Hoshi M, Pinto MR, Usui N. Purification and characterization of a vitelline coat lysin from Ciona intestinalis spermatozoa. Mol Repro Dev. 1992;32:383–388. doi: 10.1002/mrd.1080320412. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nishikawa T, Bird A. Genomic approaches reveal unexpected difference within Ciona intestinalis. J Mol Evol. 2005;61:627–635. doi: 10.1007/s00239-005-0009-3. [DOI] [PubMed] [Google Scholar]
- Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F. The evolution of house mice. Ann Rev Ecol Syst. 1993;24:119–152. doi: 10.1146/annurev.es.24.110193.001003. [DOI] [Google Scholar]
- Vacquier VD, Moy GW. Isolation of bindin: the protein responsible for adhesion of sperm to sea urchin eggs. Proc Natl Acad Sci. 1977;74:2456–2460. doi: 10.1073/pnas.74.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios HA. Reproductive isolation between species of sea urchins. Bull Mar Sci. 2007;81:191–208. [Google Scholar]
- Lee Y H, Ota T, Vacquier VD. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ, Vacquier VD. Maximum-Likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Conover WJ. Practical Nonparametric Statistics. 3. New York; Wiley Publishing; 1999. [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality. Biometrika. 1965;52:591–599. [Google Scholar]
- Wilson DJ, McVean G. Estimating diversifying selection and functional constraint in the presence of recombination. Genetics. 2006;172:1411–1425. doi: 10.1534/genetics.105.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:14005–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]