Fig. 10.
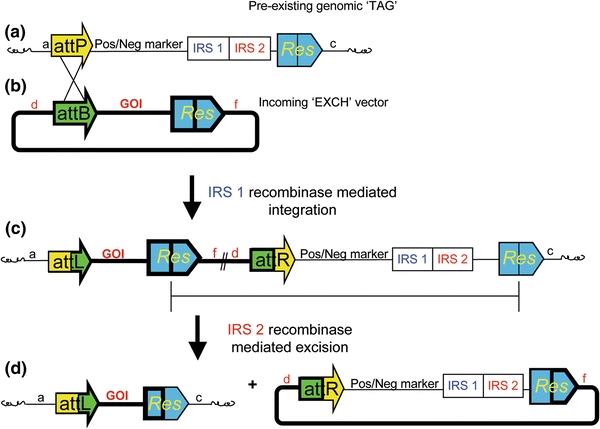
Schematic representation of the irreversible recombinase-mediated cassette exchange. This strategy simultaneously uses two irreversible recombinases, one for integration into the pre-existing chromosomal ‘TAG,’ and one for excision of unwanted DNA. This strategy traps the GOI from the incoming ‘EXCH’ cassette in the genome (seen as circle with thick lines). Even though two recombinase-mediated events are necessary, these reactions occur in a single transformation experiment. Integration of the ‘EXCH’ cassette is identified via negative selection against the presence of the Pos/Neg selectable marker. The design makes the proper alignment of the recognition sites for excision dependent on the site-specific integration event. No site-specific or random integration event where the negative selection marker is present survives negative selection. a The ‘TAG’ contains a positive–negative selectable marker and two irreversible recombinase genes flanked by the recognition sites (attP and Res) of both IRR systems, represented by yellow and blue arrows, respectively. b The ‘EXCH’ vector is a simple vector containing a GOI flanked by the recognition sites of the two corresponding IRR systems (attB and Res, represented by green and blue arrows, respectively. c Irreversible recombinase-mediated integration inserts the entire plasmid into the pre-existing ‘TAG’ loci (i.e. the yellow and green arrows are recombined—integration). Site-specific integration aligns the Res recognition sites in direct orientation (blue arrows) enabling excision. Irreversible recombinase-mediated excision removes the positive–negative marker and recombinase genes from the genome (i.e. the Res sites are recombined—excising the undesirable DNA). d Upon completion of both irreversible recombination events, the GOI has been site-specifically inserted into the genome free of unwanted plasmid backbone, marker and recombinase genes. Circularized DNA does not replicate and cannot be reintegrated into the genome via the recombinase genes as neither the attL, attR (seen as bicolored yellow and green arrows) or Res recognition sites (blue arrows) can mediate genomic integration
