Fig. 9.
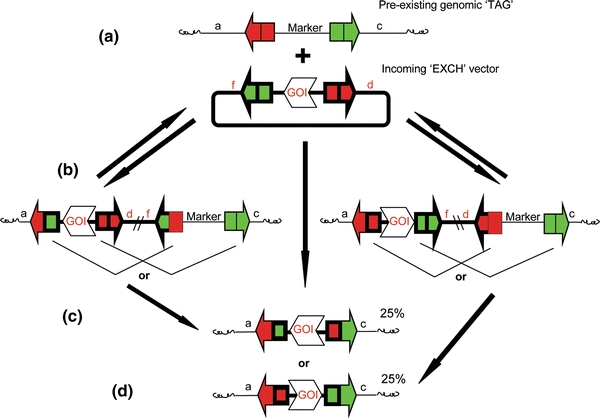
General RMCE schematic for a single bidirectional recombinase (the green and red arrows represent identical recognition sites and colors are only intended to allow visualizing of the various recombination possibilities). This technique has been shown to be an efficient way to produce transgenics with a minimum of excess DNA added to the host. This strategy combines both site-specific integration and excision mediated by one recombinase. The approach involves a targeted integration event followed by a recombinase-mediated excision event removing the unwanted transgenic DNA. Specifically, RMCE is a technique where DNA can be integrated in a specific manner to a pre-existing genomic target ‘TAG’ with a minimum of backbone DNA. The pre-existing ‘TAG’ contains a selectable marker (Marker) and is flanked by inverted RRS recognition sites (red and green arrows, thin lines). The incoming plasmid DNA contains a gene of interest (GOI) also flanked by inverted RRS recognition sites (red and green arrows, thick lines) and is termed the exchange ‘EXCH’ cassette. The recombinase will integrate the incoming ‘EXCH’ DNA into the ‘TAG’ utilizing one of the two flanking recognition sites. To proceed forward, the second set of recognition sites are then used for site-specific excision of the intervening DNA. This results in the switch the GOI of the ‘EXCH’ cassette for the (Marker) of the ‘TAG’ DNA. Although useful, inverted sites also result in ‘exchanged’ cassettes with both forward and reverse orientations making the molecular conformation more complicated and differing transgene expression levels. Not all possible conformations are represented. The recombinase can be provided in cis or in trans—not shown
