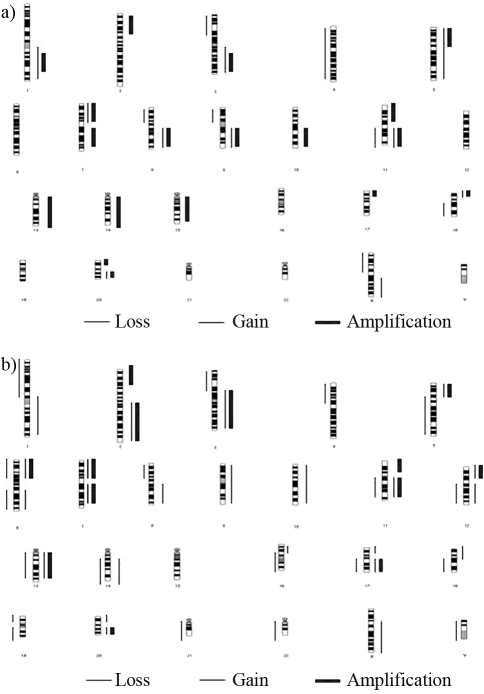
Figure 1.
Summary of copy number alterations in esophageal squamous cell carcinomas (ESCC) analyzed by comparative genomic hybridization. Regions with copy number gains are represented by thin lines on the right side of the chromosome idiograms and amplifications are represented by thick bars on the same side; regions of loss are represented by thin lines on the left side of the idiograms. (A) In ESCC cell lines, the most frequent genomic gains were observed in chromosomes 1, 2q, 3q, 5p, 6p, 7, 8q, 9q, 11q, 12p, 14, 15, 16, 17, 18p, 19q, 20q, 22q and X. Focal amplifications were found at 1q32, 2p16-22, 3q25-28, 5p13-15.3, 7p12-22, 7q21-22, 8q23-24.2, 9q34, 10q21, 11p11.2, 11q13, 13q32, 14q13-14, 14q21, 14q31-32, 15q22-26, 17p11.2, 18p11.2-11.3 and 20p11.2. Recurrent losses occurred at 3p, 4, 5q, 6q, 7q, 8p, 9, 10p, 12p, 13, 14p, 15p, 18, 20, 22, Xp and Y. (B) In clinical specimens, chromosomal gains were common in 1q, 2q, 3q, 5p, 6, 7, 8q, 9, 10, 11q, 12, 13q, 14q, 16p, 17, 18q, 20 and Xq, specifically in the regions 1p34, 2p24, 2q24-34, 3q22-ter, 7p12-22, 8q13-qter, 11p11.2, 11q13, 12p11.2, 13q21-34, 17q12, 20q12-13 and Xq27-28. Recurrent losses occurred at 1p, 3q, 4p, 5q, 6, 8p, 11q, 12q, 13q, 14q, 16q, 17q, 18q, 19, 21q, 22q and Y.