Abstract
Genetic variation was estimated in ten samples populations of Aedes aegypti from the Brazilian Amazon, by using a 380 bp fragment of the mitochocondrial NADH dehydrogenase subunit 4 (ND4) gene. A total of 123 individuals were analyzed, whereby 13 haplotypes were found. Mean genetic diversity was slightly high (h = 0.666 ± 0.029; π = 0.0115 ± 0.0010). Two AMOVA analyses indicated that most of the variation (~70%-72%) occurred within populations. The variation found among and between populations within the groups disclosed lower, but even so, highly significant values. FST values were not significant in most of the comparisons, except for the samples from Pacaraima and Rio Branco. The isolation by distance (IBD) model was not significant (r = 0.2880; p = 0.097) when the samples from Pacaraima and Rio Branco were excluded from the analyses, this indicating that genetic distance is not related to geographic distance. This result may be explained either by passive dispersal patterns (via human migrations and commercial exchange) or be due to the recent expansion of this mosquito in the Brazilian Amazon. Phylogenetic relationship analysis showed two genetically distinct groups (lineages) within the Brazilian Amazon, each sharing haplotypes with populations from West Africa and Asia.
Keywords: Aedes aegypti, dengue vector, genetic structure, gene flow, Brazilian Amazon, genetic lineages
Introduction
Aedes aegypti is the main vector of urban yellow fever and four dengue virus serotypes (DENV-1 to DENV-4). This mosquito is also involved in the transmission of other arboviruses and filarial helminthes which affect humans and several other animal species (Forattini, 2002). However, the main epidemiological problem is related to the transmission of dengue, especially in its more severe form, dengue hemorrhagic fever (DHF) (Gubler, 1998). It is estimated that worldwide 50-100 million cases of dengue fever (DF) occur every year, with 500,000 cases of DHF and at least 22,000 deaths, mainly among children (WHO, 2007). Until now, no vaccine against dengue is available, the efforts to curb the progress of this disease being based solely on vector control measures alone (Gubler, 1998).
Aedes aegypti is an urban species, being well adapted to live in close association with humans and demonstrating great adaptive capacity to the most varied environments (Paupy et al., 2000; Donalísio and Glasser, 2002). Studies have shown that ecological variation, human intervention, dispersal patterns and the constant use of insecticides may affect the genetic population structure of this vector (Bosio et al., 1998; Yan et al., 1998; Huber et al., 2002; Paupy et al., 2005; Scarpassa et al., 2008), this having been associated with heterogeneous patterns of vector competence in the transmission of dengue and urban yellow fever viruses (Failloux et al., 2002; Lourenço-de-Oliveira et al., 2002, 2004).
In 1955, Ae. aegypti came to be considered as having been eradicated from Brazil (Consoli and Lourenço-de-Oliveira, 1994). Nevertheless, a few years later it was probably re-introduced into the country through the states of Pará (1967), Bahia (1976) and Rio de Janeiro (1977) (Lourenço-de-Oliveira et al., 2004). In 1998, this vector was already present countrywide (Figueiredo, 2003; MS, 2007). In spite of vector control programs, dengue outbreaks are common, with a significant increase in cases of DHF (MS, 2007).
The Amazon region possesses ideal characteristics favoring rapid development of the life cycle, proliferation and longevity of Ae. aegypti, namely in the form of towns with large harbors, the tropical climate (high temperatures and humidity), pronounced rainy seasons and complex environmental and social factors. Currently, this vector is found in virtually all the towns of the Brazilian Amazon (SVS, 2006), its dispersion possibly having occurred via river traffic, a major travel route for persons and commercial trade between many localities of the region. Thus, studies in population genetics of these Ae. aegypti populations could provide knowledge on gene-flow patterns and colonization events, which would be important parameters for outlining new strategies for local and regional vector control.
Among the mitochondrial genes, the NADH dehydrogenase subunit 4 (ND4) gene has shown to be an excellent marker for analyzing the genetic population structure and colonization events in Ae. aegypti (Gorrochotegui-Escalante et al., 2002; Bosio et al., 2005; Costa-da-Silva et al., 2005; Herrera et al., 2006; Bracco et al., 2007; Paduan and Ribolla, 2008; Urdaneta-Marquez et al., 2008).
In this study we investigated the genetic variability of wild Ae. aegypti populations from seven towns of the Brazilian Amazon, through the use of sequences of the ND4 gene. We also analyzed four suburban neighborhoods within the city of Manaus, in order to establish the gene flow pattern at the micro-geographic level.
Materials and Methods
Collection of the mosquito
Ae. aegypti samples were collected in seven towns of the Brazilian Amazon: Belém and Santarém (state of Pará), Boa Vista and Pacaraima (state of Roraima), Rio Branco (state of Acre), and Coari and Manaus (state of Amazonas). With a view to micro-geographic analysis, individuals from four suburban neighborhoods of Manaus were sampled: Coroado, Praça 14 de Janeiro, Compensa, and Tancredo Neves (Figure 1). Information on localities, states and geographic coordinates is presented in Table 1. A total of ten samples were analyzed.
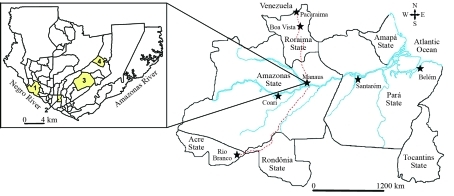
Figure 1.
Collection sites of Aedes aegypti in the Brazilian Amazon. On the larger map, rivers are represented in blue, whereas highways are represented as red dashed lines. The stars indicate collection sites. On the smaller map, the four urban neighborhoods of Manaus city are represented in yellow: 1 = Compensa; 2 = Praça 14 de Janeiro; 3 = Coroado; 4 = Tancredo Neves.
Table 1.
Localization of the Aedes aegypti populations from the Brazilian Amazon.
| State | Locality | Location code | Coordinates | Sample size |
| Amazonas | Coroado* | Cor | 3° 05' S, 59° 58' W | 19 |
| Amazonas | Praça 14 de Janeiro* | Pra | 3° 07' S, 60° 00' W | 11 |
| Amazonas | Compensa* | Com | 3° 06' S, 60° 03' W | 10 |
| Amazonas | Tancredo Neves* | TaN | 3° 03' S, 59° 56' W | 10 |
| Amazonas | Coari | Coa | 4° 05' S, 63° 08' W | 12 |
| Pará | Santarém | Stm | 2° 26' S, 54° 43' W | 14 |
| Pará | Belém | Blm | 1° 25' S, 48° 27' W | 11 |
| Roraima | Boa Vista | BoV | 2° 48' N, 60° 42' W | 12 |
| Roraima | Pacaraima | Pac | 4° 25' N, 61° 08' W | 12 |
| Acre | Rio Branco | RBr | 9° 58' S, 67° 49' W | 12 |
*neighborhoods of Manaus.
Larval and pupal stages were collected from 15 to 30 diverse artificial recipients in each town, this including the four suburbs of Manaus. The specimens collected from each recipient-site were transfered to Laboratory of Population Genetics and Evolution of Malaria and Dengue Vectors in the National Institute for Amazonian Research, in Manaus, Amazonas, Brazil. Larvae and pupae were reared to adulthood in the laboratory. Subsequently, specimens were identified by means of a key developed by Forattini (2002), to be subsequently frozen at -80 °C until genomic DNA extraction. One to two individuals from each artificial recipient were used in this study.
DNA extraction, and amplification and sequencing of the ND4 gene
Genomic DNA was extracted individually from 4th instar larvae and/or adults by using the protocol developed by Collins et al. (1987). A 380 bp fragment was amplified with the aid of primers described by Gorrochotegui-Escalante et al. (2002), under amplification conditions as outlined by Bosio et al. (2005). A negative control was used in all PCR reactions. PCR products were deposited on 1% agarose gels, stained with ethidium bromide and analyzed under UV light. These were then purified using GFX PCR DNA and a Gel Band Purification Kit (GE Healthcare, UK) according to manufacturer's recommendations. Subsequently, they were sequenced in an automated MegaBACE 1000 Analysis System sequencer (GE Healthcare, UK). All individuals were sequenced in both directions.
Statistical analysis
Sequences were edited and aligned by means of the BioEdit (Hall, 1999) and Chromas (Griffith University, Queensland, Australia) programs. Haplotype genealogy based on the parsimony method was generated through the TCS program (Clement et al., 2000). The genetic diversity of the haplotypes (h) and nucleotides (π), the number of variable sites (NV) and the average number of nucleotide differences (K), were calculated by using the DnaSP program (Rozas et al., 2003). Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997) neutrality tests, as well as haplotype (h) and nucleotide (π) diversities, the average number of nucleotide differences (K) and sequence divergence (D) calculated between the two haplotype groups, were also obtained through the DnaSP program.
Estimates of genetic distance and gene flow based on FST and Nm values, respectively, as well as hierarchical analysis (AMOVA), were calculated using the Arlequin program (Excoffier et al., 2006). FST statistics (Wright, 1921) was used to estimate the genetic structure of the populations. The number of migrants per generation (Nm), which provides an estimate of gene flow between sub-populations, was obtained through FST values. The correlation between genetic (FST) and geographic (km) distances was estimated by using Mantel's test (Mantel, 1967). The isolation by distance (IBD) hypothesis was tested with the IBDWS program (Jensen et al., 2005). Geographic distances for this analysis were obtained from the GPS and those in the Google Earth program. Levels of significance were adjusted by the Bonferroni correction whenever there were multiple tests (Rice, 1989).
Phylogenetic relationships among haplotypes were estimated by using the Mega program (Kumar et al., 2004), based on the neighbor-joining (NJ) algorithm within the Tamura-Nei genetic distance model. Bootstrap support was calculated by means of 1000 replicates. In this analysis, Aedes albopictus (GenBank accession number: EF153761) and Anopheles marajoara (GenBank accession number: AY846347) were employed as outgroups.
The haplotypes in this study were compared with those available in previous inquiries, from Mexico - AF334841-AF334865 (Gorrochotegui-Escalante et al., 2002), Peru - DQ177153-DQ177155 (Costa-da-Silva et al., 2005), and the Americas, Africa and Asia - DQ176828-DQ176831, DQ176833-DQ176843 and DQ176845-DQ176849 (Bracco et al., 2007). Only shared haplotypes are shown in Table 2. Those detected in Thailand (Bosio et al., 2005) and Venezuela (Herrera et al., 2006; Urdaneta-Marquez et al., 2008) have not yet been deposited in the GenBank, thereby precluding comparison with haplotypes in this study. The haplotypes observed in Brazil by Paduan and Ribolla (2008) (AY906835-AY906853) were not compared with those of the present study, since there was no indication of origin. The haplotypes of this study are deposited in the GenBank under accession numbers EU650405-EU650417.
Table 2.
Haplotype frequencies in the ten Aedes aegypti samples, and haplotypes shared by this study and three previous studies that used the ND4 gene.
| Haplotypes | Localities
|
Total | ||||||||||||
| Cor | Pra | Com | TaN | Coa | Stm | Blm | BoV | Pac | RBr | Mex1 | Per2 | AAA3 | ||
| H1 | 7 | 3 | 7 | 8 | 11 | 10 | 12 | 58 | H3 | H15 | ||||
| H2 | 1 | 1 | ||||||||||||
| H3 | 1 | 1 | H5 | H13 | ||||||||||
| H4 | 1 | 1 | ||||||||||||
| H5 | 1 | 1 | H3 | H1 | H16 | |||||||||
| H6 | 7 | 5 | 2 | 5 | 3 | 2 | 4 | 12 | 40 | H17 | ||||
| H7 | 1 | 1 | ||||||||||||
| H8 | 1 | 1 | ||||||||||||
| H9 | 1 | 1 | ||||||||||||
| H10 | 4 | 4 | ||||||||||||
| H11 | 3 | 2 | 5 | 2 | 12 | H6 | H7 | |||||||
| H12 | 1 | 1 | ||||||||||||
| H13 | 1 | 1 | ||||||||||||
| Total | 19 | 11 | 10 | 10 | 12 | 14 | 11 | 12 | 12 | 12 | 123 | |||
1 = haplotypes from Mexico (Gorrochotegui-Escalante et al., 2002); 2 = haplotypes from Peru (Costa-da-Silva et al., 2005); 3 = haplotypes from the Americas-Africa-Asia (Bracco et al., 2007).
Results
Distribution and frequency of haplotypes
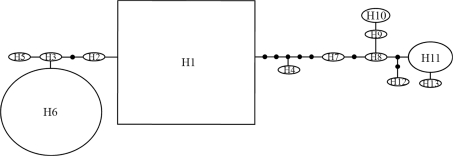
13 haplotypes were discovered from among 123 sequenced individuals of Ae. aegypti (Table 2), haplotype 1 being the most frequent (47.15%) followed by haplotype 6 (32.52%). Except for Boa Vista and Pacaraima, the latter was detected in all populations. Haplotype 11, the third most frequent haplotype, was observed in three suburbs of Manaus and in Boa Vista. Samples from Belém revealed the highest number of exclusive haplotypes (H3, H8, H9 and H10), among which H10 was observed in four individuals, whereas H3, H8 and H9 were detected in only one single individual each. Haplotype 1, represented by a rectangle, is probably the oldest (Clement et al., 2000), being separated from H6 and H11 by three and nine mutational steps, respectively (Figure 2).
Figure 2.
Haplotype network observed in the ten samples of Aedes aegypti. The size of the ellipse is proportional to the number of individuals found for each haplotype. Black circles indicate mutational steps between haplotypes.
Genetic variability and gene flow among the populations
The highest levels of both haplotype and nucleotide diversity were detected in the samples from Belém and two suburbs of Manaus (Coroado and Praça 14 de Janeiro) (Table 3). The samples from Tancredo Neves (in Manaus) displayed the highest nucleotide diversity, whereas there was no nucleotide variation in those from Pacaraima (H1) and Rio Branco (H6), thus indicating these to be monomorphic. Tajima's D and Fu's Fs neutrality tests showed no significant results (p > 0.05) in all the samples, this suggesting that these populations are in genetic equilibrium (Fu, 1997).
Table 3.
Genetic variability and neutrality tests of the ten Aedes aegypti samples from the Brazilian Amazon.
| Populations | NV | K | h | π | Tajima's D test | Fu's Fs test |
| Cor | 13 | 5.192 | 0.737 ± 0.062 | 0.0136 ± 0.0022 | 1.456 | 3.996 |
| Pra | 13 | 5.891 | 0.745 ± 0.098 | 0.0155 ± 0.0030 | 1.432 | 3.923 |
| Com | 5 | 1.955 | 0.511 ± 0.164 | 0.0051 ± 0.0015 | 0.427 | 1.926 |
| TaN | 12 | 6.666 | 0.556 ± 0.075 | 0.0175 ± 0.0023 | 2.581 | 9.078 |
| Coa | 4 | 1.712 | 0.530 ± 0.136 | 0.0045 ± 0.0013 | 1.029 | 1.900 |
| Stm | 9 | 1.868 | 0.385 ± 0.149 | 0.0049 ± 0.0021 | -1.304 | 2.386 |
| Blm | 12 | 6.472 | 0.782 ± 0.093 | 0.0170 ± 0.0020 | 2.518 | 2.672 |
| Bov | 10 | 3.030 | 0.303 ± 0.147 | 0.0079 ± 0.0038 | -0.349 | 5.937 |
| Pac | 0 | 0.000 | 0.000 ± 0.000 | 0.0000 ± 0.0000 | NC | NC |
| RBr | 0 | 0.000 | 0.000 ± 0.000 | 0.0000 ± 0.0000 | NC | NC |
| Mean | 16 | 4.399 | 0.666 ± 0.029 | 0.0115 ± 0.0010 | 1.309 | 1.418 |
NV = number of variable sites; K = average number of nucleotide differences; h = haplotype diversity; π = nucleotide diversity; NC = not calculated. Significance level for neutrality tests: p > 0.05.
Two analyses levels were used in the AMOVA test to verify the origin of genetic variation in the different hierarchical levels and groups. In the first, all the populations analyzed were considered as constituting a single group (island model), the highest variation occurring within the populations themselves (72.69%). Nevertheless, the percentage of variation was lower among samples (27.31%), even though highly significant (FST = 0.273; p < 10-5). In the second AMOVA test, the populations were grouped according to their respective states, the Amazonas group, comprising the four suburbs of Manaus (Coroado, Praça 14 de Janeiro, Compensa and Tancredo Neves) and Coari, the Pará group, comprising Santarém and Belém, the Roraima group with Boa Vista and Pacaraima, and the Acre group with Rio Branco. Similar to the first AMOVA analysis, the greater part of variation (70.66%) occurred within the populations themselves, with a highly significant value (FST = 0.293, p < 10-5). The percentage of variation between populations within the states (18.87%) was also highly significant (FSC = 0.210, p < 10-5), whereas among the states, the percentage of variation (10.47%) was low and insignificant (FCT = 0.104; p > 0.01) (data not shown).
Significant FST values (p < 0.001) were observed, after Bonferroni correction, in most of the comparisons involving samples from Rio Branco, with the corresponding Nm values varying from 0.0 to 1.0, thereby indicating from an absent to a reduced gene flow (Table 4). FST values were high in the six comparisons involving samples from Pacaraima, three of which becoming statistically significant after the Bonferroni correction. There were no statistically significant comparisons among samples from the four suburbs of Manaus, although a high FST value (0.366; Nm = 0.9) was obtained between samples from Compensa and Tancredo Neves, which was not significant after Bonferroni correction. This could be explained on considering that in Compensa only group II haplotypes were found, whereas in the samples from Tancredo Neves, haplotypes of two groups in similar frequencies were sampled.
Table 4.
Genetic distances (FST values) and effective number of migrants (Nm), above and below the diagonal, respectively, among samples of Aedes aegypti from the Brazilian Amazon.
| Population | Cor | Pra | Com | TaN | Coa | Stm | Blm | BoV | Pac | RBr |
| Cor | - | -0.061 | 0.055 | 0.085 | 0.073 | 0.084 | 0.147 | 0.066 | 0.228 | 0.330* |
| Pra | Inf. (5) |
- | 0.120 | 0.000 | 0.146 | 0.166 | 0.087 | 0.127 | 0.353* | 0.325 |
| Com | 8.5 (8) |
3.7 (4) |
- | 0.366 | -0.089 | -0.037 | 0.373 | 0.094 | 0.214 | 0.699* |
| TaN | 5.4 (5) |
Inf. (10) |
0.9 (13) |
- | 0.398 | 0.403 | 0.037 | 0.302 | 0.554* | 0.476 |
| Coa | 6.4 (368) |
2.9 (363) |
Inf. (360) |
0.8 (373) |
- | -0.009 | 0.405 | 0.111 | 0.210 | 0.706* |
| Stm | 5.5 (589) |
2.5 (593) |
Inf. (597) |
0.7 (585) |
Inf. (953) |
- | 0.402* | 0.014 | 0.051 | 0.732* |
| Blm | 2.9 (1288) |
5.3 (1293) |
0.8 (1297) |
13.1 (1284) |
0.7 (1654) |
0.7 (702) |
- | 0.323 | 0.545* | 0.506 |
| BoV | 7.0 (656) |
3.4 (660) |
4.8 (658) |
1.2 (654) |
4.0 (812) |
35.1 (880) |
1.0 (1435) |
- | 0.010 | 0.716 |
| Pac | 1.7 (840) |
0.9 (843) |
1.8 (841) |
0.4 (837) |
1.9 (967) |
9.3 (1040) |
0.4 (1550) |
5.0 (185) |
- | 1.000* |
| RBr | 1.0 (1153) |
1.0 (1148) |
0.2 (1146) |
0.6 (1158) |
0.2 (827) |
0.2 (1670) |
0.5 (2339) |
0.2 (1618) |
0.0 (1756) |
- |
*p < 0.001, after Bonferroni correction; Inf. = infinity; geographic distances (km) are in parentheses.
The correlation between genetic and geographic distances was statistically significant (r = 0.5815; p = 0.006) when taking all the populations into consideration, thereby suggesting isolation by distance (IBD). However, when the samples from Pacaraima and Rio Branco, the most genetically structured, were removed from the analysis, correlation was non-significant (r = 0.2880; p = 0.097), thus indicating that gene flow is not related to geographic distance.
Phylogenetic analysis
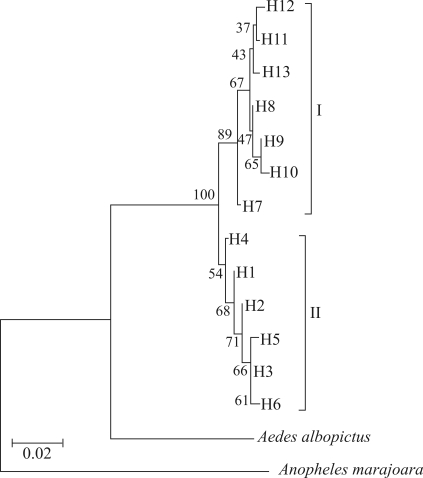
Phylogenetic relationships among the 13 haplotypes recovered two groups, with a bootstrap support of 89% between them (Figure 3). Group I was composed of seven haplotypes, the third most frequent (H11), H10 (from Belém) and five unique haplotypes (H7, H8, H9, H12 and H13). In this group, haplotype H7 observed in Santarém, was the most distant from the remainder. Furthermore, there were tree subgroups in group I: 1) cluster H11, H12 and H13, composed of samples from Manaus and Boa Vista; 2) cluster H8, H9 and H10, comprising the sample from Belém; and 3) H7, consisting of the sample from Santarém. However, bootstrap support values for splits leading to these groups were low. Group II consisted of six haplotypes, the two most common and widespread (H1 and H6) and four unique haplotypes (H2, H3, H4 and H5).
Figure 3.
Phylogenetic relationships among Aedes aegypti haplotypes, based on the neighbor-joining (NJ) algorithm under the Tamura-Nei genetic distance model. Bootstrap values are marked on the branches.
Haplotype diversity within groups I and II was 0.6571 and 0.5276, respectively, whereas nucleotide diversity was 0.00590 and 0.00325, respectively. The average number of nucleotide differences (K) between groups I and II was 10.308, whereas nucleotide divergence (D) was 0.02713.
Discussion
Distribution and frequency of haplotypes
In this study, although H1 manifested the highest frequencies in the samples from Pacaraima (100%), Boa Vista (83%) and Santarém (79%), it was totally absent in those from Tancredo Neves, Belém and Rio Branco. These findings suggest that gene flow is absent between Pacaraima and Rio Branco, and reduced between Pacaraima and Belém, and Boa Vista and Rio Branco (see Table 4). H1 was found by Costa-da-Silva et al. (2005) in Peru (as H3), and by Bracco et al. (2007) (as H15) in the localities of Boa Vista, Manaus, Cariacica (state of Espirito Santo), Santos (state of São Paulo), and Piura (Peru), but was not observed in Mexico (Gorrochotegui-Escalante et al., 2002). Nevertheless, the haplotypes in this study were not compared with haplotypes from Venezuela since the latter were not available (Herrera et al., 2006). Based on these results, we were unable to propose an origin for H1.
On the contrary, H6 was absent in the samples from Pacaraima and Boa Vista, but was detected in all the others in this study. In an extensive study on the Americas, Africa and Asia, Bracco et al. (2007) observed that H6 (as H17) was exclusive to Brazil, with a wide distribution. H6 has not been detected neither in Mexico (Gorrochotegui-Escalante et al., 2002) nor Peru (Costa-da-Silva et al., 2005). It is our opinion that H6 may either have arisen in Brazil or was introduced from other, as yet unstudied, regions of the world. Additional inquiries may clarify the origin of this haplotype. H11, observed in Boa Vista and three suburbs in Manaus, was also discovered in Mexico as H6 (Gorrochotegui-Escalante et al., 2002), and in other locations in Brazil (the towns of Boa Vista and Potim), besides the U.S.A. and Senegal (West Africa) as H7 (Bracco et al., 2007). This haplotype may have arisen in West Africa, to later spread to other regions in the world. Thus, H11 may represent a further introduction into Brazil. H3, observed in only one individual from Belém, was also found in Mexico as H5 (Gorrochotegui-Escalante et al., 2002) and Asia as H13 (Bracco et al., 2007). This haplotype may have been introduced into the Americas (including Brazil) from Asia. H8, H9 and H10, exclusive of Belém, have no shared with any other haplotype from previous studies. These haplotypes may represent recent mutations or introductions that have not yet spread.
Genetic variability and gene flow among the populations
In this study, mean genetic diversity proved to be relatively high, whereas both Costa-da-Silva et al. (2005), on studying three populations from Peru, and Bosio et al. (2005), when analyzing 19 populations from Thailand, estimated a lower value (π = 0.0079) than that presented herein. The highest values of nucleotide diversity were found in populations from Mexico (π = 0.0143; Gorrrochotegui-Escalante et al., 2002), Venezuela (π = 0.0187; Herrera et al., 2006), Brazil (π = 0.0174; Paduan and Ribolla, 2008) and the Americas-Africa-Asia (π = 0.0199; Bracco et al., 2007). Comparing our π values to those of Venezuela, a lower haplotype number (7) was observed in the latter, whereas π values were higher, this possibly indicating that the Venezuelan populations are older than those of the Brazilian Amazon (Kambhampati and Rai, 1991). However, these findings could be the outcome of mosquito control efforts, with the consequential loss of intermediate haplotypic lineages, thus resulting in high divergence among haplotypes.
The highest level of genetic diversity found in the samples from Belém and suburbs of Manaus (Coroado, Praça 14 de Janeiro and Tancredo Neves) may be coincident with demographic density being the highest in these two port-towns in north Brazil, besides their both being situated on the banks of two large rivers (Amazon and Negro), thereby being exposed to an intense flow of both persons and trade. Furthermore, the industrial center of Manaus is one of the largest in Brazil. These factors are likely to favor multiple introductions and/or the dispersal of Ae. aegypti, thus contributing to an increase in gene flow between populations, with the subsequent rise in genetic variation. The four exclusive haplotypes found in Belém may support this hypothesis. These findings could imply that, despite the constant use of insecticides for vector control, these Ae. aegypti populations may have a large effective population size.
The two AMOVA analyses indicated that a greater part of the variation occurred within populations (~70%-72%), which can probably be attributed to the presence of two genetically distinct sympatric haplotype lineages. The significant variation (27.31%) among populations (non-grouped) could be primarily due to the high genetic differentiation found between Pacaraima and Rio Branco, whereas the significant variation (18.87%) between populations within the states themselves could result mainly from the differentiation observed between Belém and Santarém (Pará group). This indicates that a significant genetic structure occurs in Ae. aegypti from the Brazilian Amazon. Similar results were found in Mexico (Gorrochotegui-Escalante et al., 2002) and Venezuela (Herrera et al., 2006).
In this study, the gene flow pattern was related to H1 and H6 frequencies. In spite of the great distance between Santarém and Boa Vista (~880 km), gene flow was extensive (Nm = 35.1) due to the high frequency of H1 in both populations. The free gene flow (Nm = 13.1) between Belém and Tancredo Neves, about 1,284 km apart, was directly related to H6 frequency. Nevertheless, this result may indicate distinct introduction events followed by the recent expansion of Ae. aegypti in the Brazilian Amazon (less than 25 years) rather than the current gene flow.
Of the ten samples analyzed herein, those from Rio Branco and Pacaraima were the most genetically structured and isolated by distance. This is mainly why genetic variation was absent in these samples, theses results possibly being consistent with two hypotheses: 1) the Rio Branco and Pacaraima Ae. aegypti populations were founded by few individuals (founder effect) and have maintained low effective population sizes; or 2) the sizes of these populations were reduced due to the constant use of insecticides, thereby generating repeated bottleneck effects and/or populations founded by few individuals through cycles of extinction and re-colonization. Both effects could imply the reduction of genetic variability through genetic drift (Hartl and Clark, 1997), thus possibly resulting in highly divergent populations.
All told, we observed higher rates of gene flow for populations of the Brazilian Amazon when compared to the two previous studies undertaken in Brazil. Ayres et al. (2003) analyzed the Ae. aegypti populations of five Brazilian states with RAPD markers, and found high levels of genetic differentiation and reduced gene flow at both the macro-geographic (Nm = 0.54) and micro-geographic (Nm = 0.69) levels. Paduan et al. (2006) also used RAPD markers in populations from six other Brazilian states and observed reduced gene flow among all of these (Nm = 0.65), even though from the same state (Nm = 0.83). Compared to the present study, these results suggest that levels of genetic variability and differentiation among Brazilian Ae. aegypti populations are relatively high. Nevertheless, most of the populations of the Brazilian Amazon analyzed here manifested higher gene flow, probably due to human migration and river or land trading movement, thereby favoring the dispersal of this vector in the region. Alternatively, this could indicate the recent expansion of Ae. aegypti throughout this area. The absence of isolation by distance, when the samples from Pacaraima and Rio Branco were removed from the analyses, supports these hypotheses. Identical results were found in samples from Mexico (Gorrochotegui-Escalante et al., 2002), Venezuela (Herrera et al., 2006) and Brazil (Scarpassa et al., 2008). Furthermore, the active dispersal patterns (flight) of Ae. aegypti have been estimated as being between 100 and 800 m (Ordonez-Gonzalez et al., 2001; Honorio et al., 2003), this dispersal mechanism obviously not being a possible explanation for our results.
Two haplotype groups were indicated through phylogenetic relationship analysis, which is in accordance with previous studies (Gorrrochotegui-Escalante et al., 2002; Bosio et al., 2005; Herrera et al., 2006; Bracco et al., 2007; Paduan and Ribolla, 2008). Group I shared haplotypes with Senegal (West Africa), whereas group II shared haplotypes with Asia. Due to the appearance of similar results with the mtDNA COI gene in most of our samples (Scarpassa et al., 2008), we believe that H1 from group II may have originated in East Africa. Even though haplotype and nucleotide diversity values in group I were higher than in group II, the latter is probably the more ancient, since it presents the oldest and most widespread haplotypes. Bracco et al. (2007) found slightly higher values for K (12.015) and D (0.03207) between the two haplotype groups than those disclosed in the present study. This could be related to the colonization history of these populations (Scarpassa et al., 2008). These results are consistent with the presence of two genetic lineages within the Brazilian Amazon, these being sympatric in Manaus, Boa Vista and Belém. Based on previous findings (Mousson et al., 2005; Bracco et al., 2007) and those arising from this study, group II (the older lineage) may have persisted independent of eradication programs in the Americas, this including in Brazil.
Concluding, the data in this study indicate multiple introductions of Ae. aegypti into the Brazilian Amazon. Cluster analysis clearly showed two genetic lineages, each of which sharing haplotypes with West Africa and Asia, thereby suggesting that Brazilian Amazon populations probably originated from these regions. The existence of distinct lineages within the Brazilian Amazon could imply differences in the susceptibility for transmitting dengue and urban yellow fever viruses (Beerntsen et al., 2000; Failloux et al., 2002; Lourenço-de-Oliveira et al., 2004; Urdaneta-Marquez et al., 2008) and in responses to vector control programs.
Acknowledgments
We thank Dr. Norma Gorrochotegui-Escalante, who kindly sent us (VMS) the primers used in this study, as well as Lúzia de Melo Mustafa and her team (Fundação de Vigilância em Saúde, Manaus, Amazonas) and the Secretaries of Health teams in Belém, Santarém, Boa Vista, Pacaraima, Coari and Rio Branco (Dulcinéia Barros de Aguiar, Agostinho Aroldo Limeira, Claudio Rodrigues de Souza) for providing facilities and logistic support during field collections. We also thank the Thematic Laboratory of Molecular Biology at INPA for access to the MegaBace sequencer and the automated sequencing analyses, and Dr. Jan E. Conn who improved an early draft of this manuscript. This research was supported by the Brazilian agency “Fundação de Amparo à Pesquisa do Estado do Amazonas” (FAPEAM) to VMS, and by the Ministry of Science and Technology/INPA, Brazil. RSLJ obtained a Master's degree through the Graduate Program in Genetics, Conservation and Evolutionary Biology at INPA, and received a fellowship from CAPES.
Footnotes
Associate Editor: Louis Bernard Klaczko
References
- Ayres C.F.J., Melo-Santos M.A.V., Solé-Cava A.M., Furtado A.F. Genetic differentiation of Aedes aegypti (Diptera, Culicidae), the major dengue vector in Brazil. J Med Entomol. 2003;40:430–435. doi: 10.1603/0022-2585-40.4.430. [DOI] [PubMed] [Google Scholar]
- Beerntsen B.T., James A.A., Christensen B.M. Genetics of mosquito vector competence. Microbiol Mol Biol Rev. 2000;64:115–137. doi: 10.1128/mmbr.64.1.115-137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio C.F., Beaty B.J., Black IVth W.C. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am J Trop Med Hyg. 1998;59:965–970. doi: 10.4269/ajtmh.1998.59.965. [DOI] [PubMed] [Google Scholar]
- Bosio C.F., Harrington L.C., Jones J.W., Sithiprasasna R., Norris D.E., Scott T.W. Genetic structure of Aedes aegypti populations in Thailand using mitochondrial DNA. Am J Trop Med Hyg. 2005;72:434–442. [PubMed] [Google Scholar]
- Bracco J.E., Capurro M.L., Lourenço-de-Oliveira R., Sallum M.A.M. Genetic variability of Aedes aegypti in the Americas using a mitochondrial gene: Evidence of multiple introductions. Mem Inst Oswaldo Cruz. 2007;102:573–580. doi: 10.1590/s0074-02762007005000062. [DOI] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K.A. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Collins F.H., Mendez M.A., Rasmussen M.O., Mehaffey P.C., Besansky N.J., Finnerty V. A ribossomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Consoli R.A.G.B., Lourenço-de-Oliveira R. Principais Mosquitos de Importância Sanitária no Brasil. Rio de Janeiro: Fiocruz; 1994. [Google Scholar]
- Costa-da-Silva A.L., Capurro M.L., Bracco J.E. Genetic lineages in the yellow fever mosquito Aedes (Stegomyia) aegypti (Diptera, Culicidae) from Peru. Mem Inst Oswaldo Cruz. 2005;100:539–544. doi: 10.1590/s0074-02762005000600007. [DOI] [PubMed] [Google Scholar]
- Donalísio M.R., Glasser C.M. Vigilância entomológica e controle de vetores do dengue. Rev Brasil Entomol. 2002;5:259–279. [Google Scholar]
- Excoffier L., Laval G., Schneider S. An Integrated Software Package for Population Genetics Data Analysis, v. 3.01. University of Berne: Computational and Molecular Population Genetics Lab, Institute of Zoology; 2006. [PMC free article] [PubMed] [Google Scholar]
- Failloux A.-.B., Vazeille M., Rodhain F. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti. J Mol Evol. 2002;55:653–663. doi: 10.1007/s00239-002-2360-y. [DOI] [PubMed] [Google Scholar]
- Figueiredo L.T.M. Dengue in Brazil: Past, present and future perspective. Dengue Bull. 2003;27:25–33. [Google Scholar]
- Forattini O.P. Culicidologia Médica. V. 2. São Paulo: Editora da Universidade de São Paulo; 2002. [Google Scholar]
- Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochotegui-Escalante N., Gomez-Machorro C., Lozano-Fuentes S., Fernandez-Salas I., Munoz M.L., Farlan-Ale J.A., Garcia-Rejon J., Beaty B.J., Black IVth W.C. Breeding structure of Aedes aegypti populations in Mexico varies by region. Am J Trop Med Hyg. 2002;66:213–222. doi: 10.4269/ajtmh.2002.66.213. [DOI] [PubMed] [Google Scholar]
- Gubler D.J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hartl D.L., Clark A.G. Principles of Population Genetics. 3rd edition. Sunderland : Sinauer Associates; 1997. [Google Scholar]
- Herrera F., Urdaneta L., Rivero J., Zoghbi N., Ruiz J., Carrasquel G., Martínez J.A., Pernalete M., Villegas P., Montoya A. Population genetic structure of the dengue mosquito Aedes aegypti in Venezuela. Mem Inst Oswaldo Cruz. 2006;101:625–633. doi: 10.1590/s0074-02762006000600008. [DOI] [PubMed] [Google Scholar]
- Honório N.A., Silva W.D., Leite P.J., Gonçalves J.M., Lounibos L.P., Lourenço-de-Oliveira R. Dispersal of Aedes aegypti and Aedes albopictus (Diptera, Culicidae) in an urban endemic dengue area in the State of Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2003;98:191–198. doi: 10.1590/s0074-02762003000200005. [DOI] [PubMed] [Google Scholar]
- Huber K., Loan L.L., Hoang T.H., Ravel S., Rodhain F., Failloux A.-.B. Genetic differentiation of the dengue vector, Aedes aegypti (Ho Chi Minh City, Vietnam) using microsatellites markers. Mol Ecol. 2002;11:1629–1635. doi: 10.1046/j.1365-294x.2002.01555.x. [DOI] [PubMed] [Google Scholar]
- Kambhampati S., Rai K.S. Mitochondrial DNA variation within and among populations of the mosquito Aedes albopictus. Genome. 1991;34:288–292. doi: 10.1139/g91-046. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lourenço-de-Oliveira R., Vazeille M., Filippis A.M.B., Failloux A.-.B. Oral susceptibility to yellow fever virus of Aedes aegypti from Brazil. Mem Inst Oswaldo Cruz. 2002;97:437–439. doi: 10.1590/s0074-02762002000300031. [DOI] [PubMed] [Google Scholar]
- Lourenço-de-Oliveira R., Vazeille M., Filippis A.M.B., Failloux A.-.B. Aedes aegypti in Brazil: Genetically differentiated populations with high susceptibility to dengue and yellow fever viruses. Trans R Soc Trop Med Hyg. 2004;98:43–54. doi: 10.1016/s0035-9203(03)00006-3. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Mousson L., Dauga C., Garrigues T., Schaffner F., Vazeille M., Failloux A.-.B. Phylogeography of Aedes (Stegomyia) aegypti (Skuse) (Diptera, Culicidae) based on mitochondrial DNA variations. Genet Res. 2005;86:1–11. doi: 10.1017/S0016672305007627. [DOI] [PubMed] [Google Scholar]
- Ordonez-Gonzalez J., Mercador-Hernandez R., Florez-Suarez A.E., Fernandez-Salas I. The use of sticky ovitraps to estimate of Aedes aegypti in Northeastern Mexico. J Am Mosq Control Assoc. 2001;17:93–97. [PubMed] [Google Scholar]
- Paduan K.S., Araújo-Júnior J.P., Ribolla P.E.M. Genetic variability in geographical populations of Aedes aegypti (Diptera, Culicidae) in Brazil elucidated by molecular markers. Genet Mol Biol. 2006;29:391–395. [Google Scholar]
- Paduan K.S., Ribolla P.E.M. Mitochondrial DNA polymorphism and heteroplasmy in population of Aedes aegypti in Brazil. J Med Entomol. 2008;45:59–67. doi: 10.1603/0022-2585(2008)45[59:mdpahi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Paupy C., Vazeille-Falcoz Mousson M., Rodhain F., Failloux A.-.B. Aedes aegypti in Tahiti and Moorea (French Polynesia): Isoenzyme differentiation in the mosquito population according to human population density. Am J Trop Med Hyg. 2000;62:217–224. doi: 10.4269/ajtmh.2000.62.217. [DOI] [PubMed] [Google Scholar]
- Paupy C., Chantha N., Reynes J.M., Failloux A.-.B. Factors influencing the population structure of Aedes aegypti from the main cities in Cambodia. Heredity. 2005;95:144–147. doi: 10.1038/sj.hdy.6800698. [DOI] [PubMed] [Google Scholar]
- Rice W.R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánches-DelBarrio J.C., Messeguer X., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Scarpassa V.M., Cardoza T.B., Cardoso-Júnior R.P. Population genetics and phylogeography of Aedes aegypti (Diptera, Culicidae) from Brazil. Am J Trop Med Hyg. 2008;78:895–903. [PubMed] [Google Scholar]
- Secretaria de Vigilancia em Saude. Boletim da Situação Epidemiológica da Dengue Semana Epidemiológica n. 52. Brasília: Ministério da Saúde; 2006. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphisms. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdaneta-Marquez L., Bosio C., Herrera F., Rubio-Palis Y., Salasek M., Black IVth W.C. Genetic relationships among Aedes aegypti collections in Venezuela as determined by mitochondrial DNA variation and nuclear single nucleotide polymorphisms. Am J Trop Med Hyg. 2008;78:479–491. [PubMed] [Google Scholar]
- Wright S. Systems of mating. Genetics. 1921;6:111–178. doi: 10.1093/genetics/6.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Chadee D.D., Severson D.W. Evidence for genetic hitchhiking effect associated with insecticide resistence in Aedes aegypti. Genetics. 1998;148:793–800. doi: 10.1093/genetics/148.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Ministerio da Saude (MS) [August 5, 2007]. Available from: http://portal.saude.gov.br/portal/arquivos/pdf/dengue1203.pdf.
- World Health Organization (WHO) [October 15, 2007]. Available from: www.who.int/csr/disease/dengue/impact/en/index.html.