Figure 3.
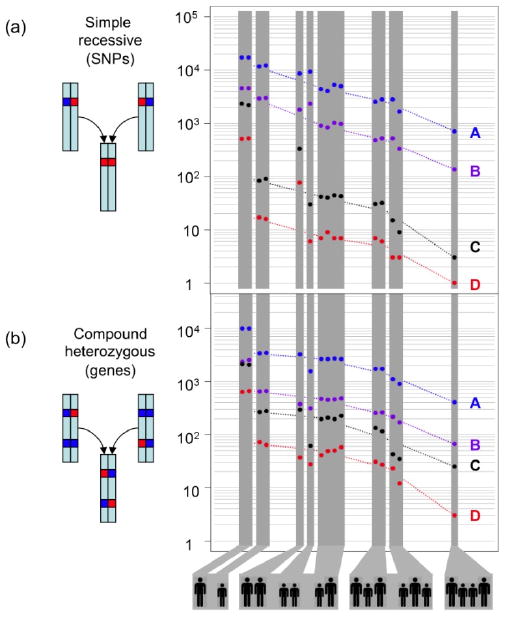
The power of family genome inheritance analysis. The number of false-positive candidates drops exponentially as the number of family members increases. (A) Number of candidate SNVs consistent with a simple recessive inheritance mode. (B) Number of candidate genes consistent with a compound heterozygous model. The different groupings of parents (large silhouettes) and children (small silhouettes) are depicted below. Dashed lines join the average values of each grouping. For this figure, probably detrimental includes missense, nonsense, splice defect, and non-initiation; possibly detrimental also includes UTR, non-coding, and splice-region. A block of SNVs such that all SNPs in the block are within 5 kb of another SNV in the block is counted only once, as together these are likely to encode at most one phenotype. A: all possibly detrimental SNVs; B: all probably detrimental SNVs; C: rare possibly detrimental SNVs; D: rare probably detrimental SNVs.
