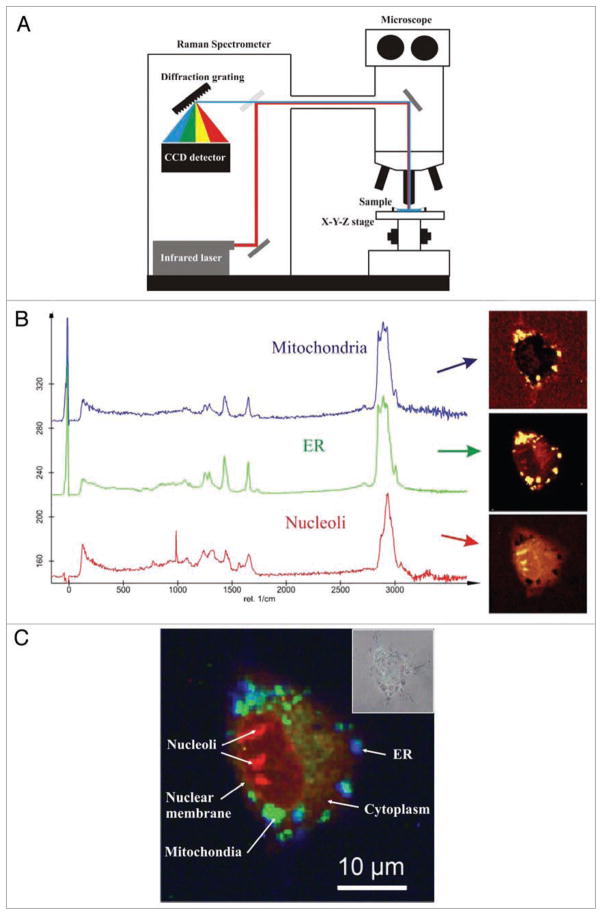
Figure 4.
Raman micro-spectroscopy in single-cell analysis of apoptosis. (A) Basic schematics of confocal Raman microscope. Scattered photons of light are collected and spectrally analyzed after passing through the diffraction grating and a CCD detector. Raman scattering occurs when photon of light collides with a molecule and interacts with its electron cloud and the chemical bonds. The inelastic interaction changes the vibration state of the molecule. Exited vibrational quantum stage of the molecule leads to an energy shift between the incident light and the back-scattered light. This energy shift is a unique chemical fingerprint of the molecule and can be used for qualitative and quantitative analysis of live cells. (B) Averaged basic Raman spectra obtained from different cellular compartments: mitochondria, endoplasmic reticulum (ER) and nucleoli, in a living HeLa cell (left). Basic spectra were constructed by averaging 10,000 spectral sets. Raman images were generated by the fit algorithm using preselected basic spectra from different regions of a cell (right). (C) Composite and pseudo-colored Raman image of a cell. Different cellular structures can be easily analyzed using spectral fingerprints. Inset shows a conventional brightfield image of the cell. Raman spectra were obtained using a WITec CRM 200 Confocal Raman Microscope using a x60 water-immersion objective and a frequency-doubled Nd:YAg laser (532 nm, 10 mW). Scan range represented region size of 100 × 100 pixels (10,000 spectra). Raman data shown are a courtesy of WITec GmbH (Ulm, Germany).