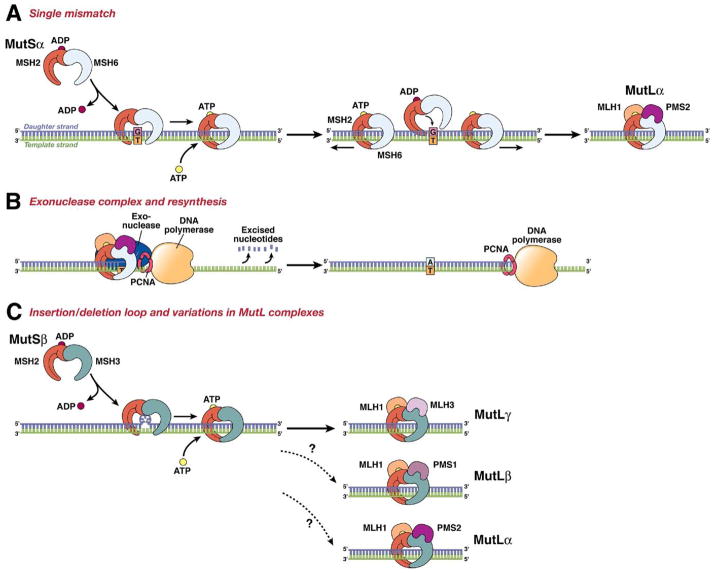
Figure 2.
The DNA MMR system functions through a series of steps. (A) MSH2–MSH6 (MutSα) recognizes single base-pair mismatches, in which the DNA polymerase has matched the wrong base (G) with the T on the template (shown on left), and creates a sliding clamp around the DNA. This step that requires the exchange of adenosine triphoshpate (ATP) for adenosine diphosphate (ADP) (by MSH2, but not MSH6 or MSH3). The complex diffuses away from the mismatch site, which is then bound by the MLH1-PMS2 (MutLα) complex (right). This “matchmaker” complex moves along the new DNA chain until it encounters the DNA polymerase complex. (B) The DNA MMR protein sliding clamp interacts with exonuclease-1, proliferating cell nuclear antigen (PCNA), and DNA polymerase. This complex excises the daughter strand back to the site of the mismatch (shown on left). Eventually, the complex falls off the DNA and resynthesis occurs, correcting the error. (C) Variations on the DNA MMR theme. Whereas MSH2–MSH6 recognizes single pair mismatches and small IDLs, MSH2–MSH3 (MutSβ) complements this by also recognizing larger IDLs (shown on left). The right side shows the possible interactions with different MutL dimers, as MLH1 can dimerize with PMS2, PMS1, or MLH3. The preferred interaction with MSH2–MSH3 is MLH1–MLH3 (MutLγ), but the precise roles of the other MutL heterodimers in this reaction are not entirely understood.