Figure 5.
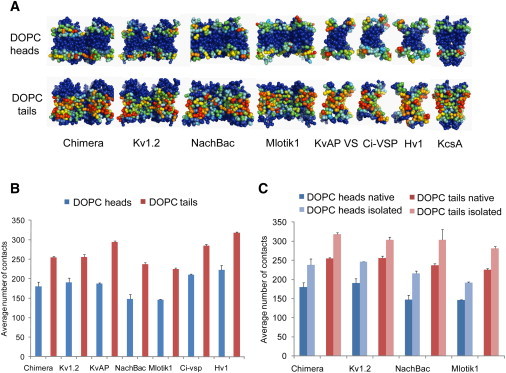
Patterns of protein lipid contacts are conserved among VS homologs. (A) Average number of contacts over the MD simulations between amino-acid side chains and DOPC head/tail groups mapped onto the channel and VS CG structures. Densities range from low contact number (blue) to high contact number (red). A cutoff distance of 6 Å was used to score a contact (as is standard for CG models). (B) Average number of contacts between the VS domains of various homologs and DOPC heads or tail groups. (C) Average number of contacts between the VS domains and DOPC heads or tail groups, comparing the VS domain simulated in isolation versus being part of the intact native channel tetramer. See also Fig. S3B.
