Figure 1.
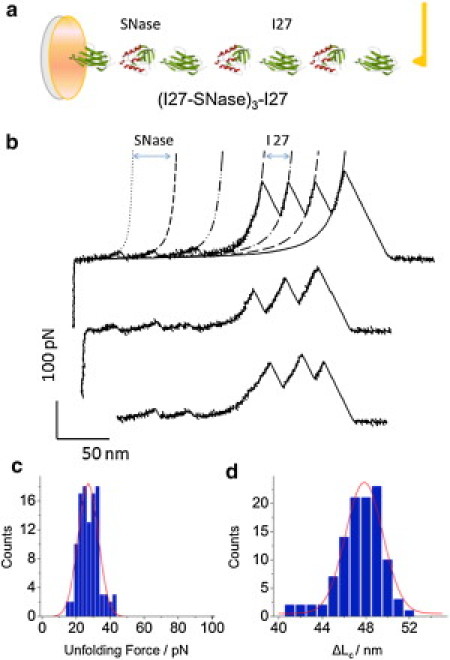
(a) Simplified structure of the chimera protein (I27-SNase)3-I27. (b) Representative unfolding force-extension traces of (I27-SNase)3-I27, the stretching speed of 200 nm/s. The AFM data are fitted with two families of WLC curves: the fits to small force peaks used the contour length increment ΔLc = 46.5 nm and the persistence length p = 0.57 nm (short dashed lines); the fits to the large force peaks used ΔLc = 28 nm and p = 0.36 nm (long dashed lines). (c) Histogram of unfolding forces of SNase domains. (Solid line) Gaussian fit to the data with 〈Funfolding〉 = 26.3 ± 0.5 pN (mean ± SE), the number of force peaks analyzed, n = 108). (d) Histogram of contour length increments ΔLc, attributed to the unfolding of SNase. (Solid line) Gaussian fit with 〈ΔLc〉 = 47.0 ± 0.2 nm (mean ± SE, n =108).
