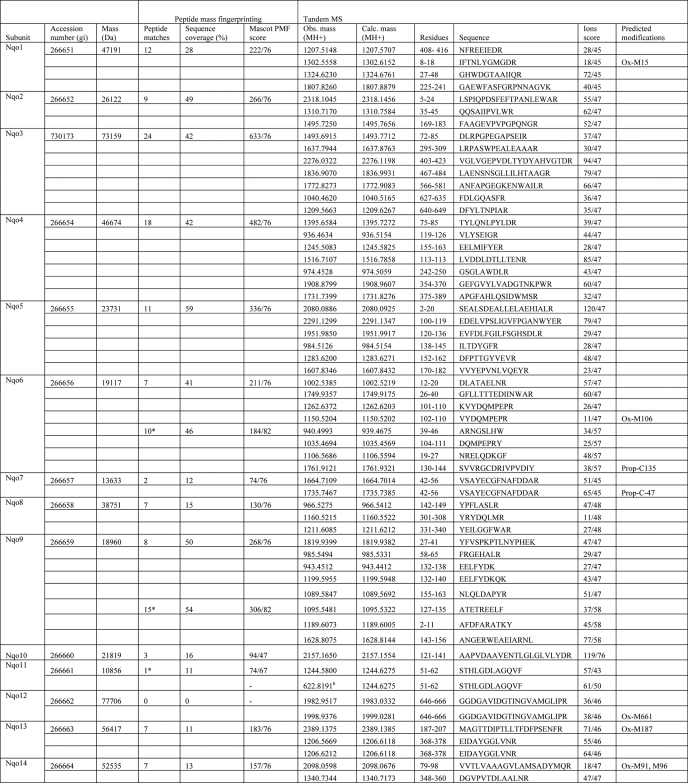
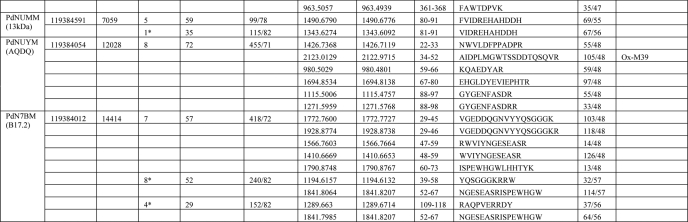
TABLE 4.
Identification of subunits of complex I by mass spectrometrya
a For peptide mass fingerprinting, data were searched against the current NCBI nonredundant nucleotide database using Mascot software (Matrix science) with a tolerance of 70 ppm. Sequences from strain ATCC 13543 were used for the calculation of the subunit mass, except for the supernumerary subunits, for which sequences are available only from strain Pd 1222. Obs. mass, observed mass; Calc. mass, calculated mass; Ox, oxidized; Prop, propionate. Samples were trypsin-digested unless the number of peptide matches is marked with an asterisk, which indicates that this sample was chymotrypsin-digested. For supernumerary subunits, the bovine homologue name is shown in brackets.
b Sample was further analyzed using the Orbitrap mass spectrometer.