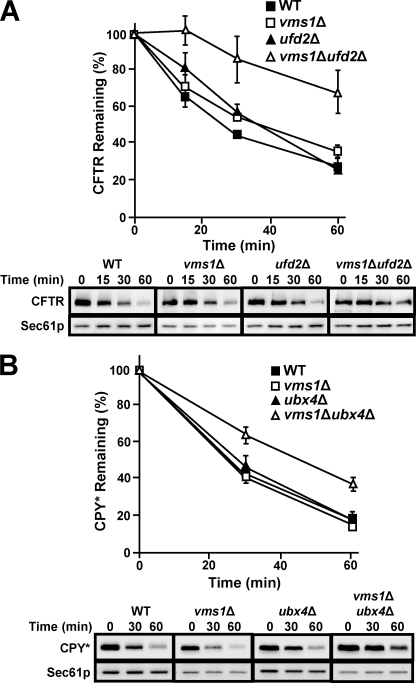
FIGURE 5.
ERAD defects are exacerbated in yeast lacking VMS1 and UFD2 or UBX4. The stability of the indicated ERAD substrate was examined by cycloheximide chase analysis as described under “Experimental Procedures” at either 40 (A) or 30 °C (B). Anti-HA antibody was used to detect each epitope-tagged ERAD substrate, and Sec61p served as a loading control. Data represent the means of three independent experiments ± S.D. (error bars). For the graphs in A and B, wild type cells are represented by black squares, and vms1Δ yeast is represented by white squares. Black triangles represent ufd2Δ in A and ubx4Δ in B, whereas white triangles represent vms1Δufd2Δ in A and vms1Δubx4Δ in B. Shown at the bottom of each panel are representative immunoblots that were used to obtain the data in each graph.