Figure 3.
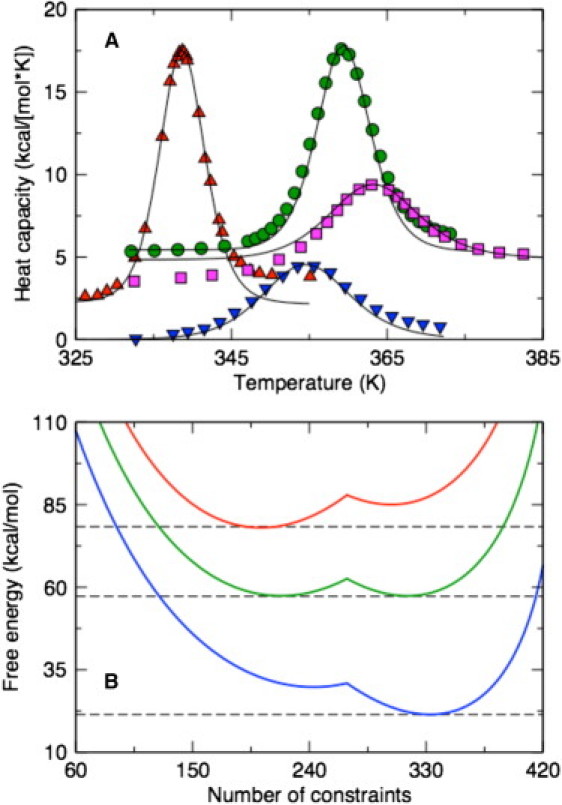
(A) The McDCM (solid lines) reproduce experimental Cp curves across a diverse set of proteins (up-triangles, lysozyme; circles, thioredoxin; squares, ubiquitin; and down-triangles, Protein G). (B) The free energy landscapes for protein G (top to bottom) at T = 310, 355, and 391 K. At Tm = 355 K, the native and disordered state basins are competitive, whereas at T1Tm, one state is clearly more probable than the other. Dashed lines are provided to guide the eye.
