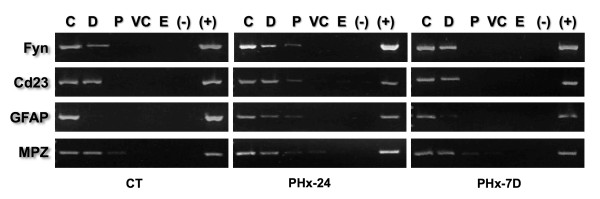
Figure 7.
Positional mapping relative to the NM of unrelated gene sequences. Nucleoids from rat hepatocytes were treated with DNase I (0.5 U/ml) for different times. The residual NM-bound DNA in the partially digested samples was directly used as template for PCR amplification of small target sequences (Table 2) located in the 5' ends of the corresponding genes (Fyn, CD23, GFAP and MPZ). Each of such genes is located in a different chromosome and thus they represent separate chromosome territories within the nucleus. The specific amplicons were resolved in 2% agarose gels and stained with ethidium bromide (0.5 μg/ml). CT, control G0 hepatocytes; PHx-24 h, hepatocytes 24 h after partial hepatectomy; PHx-7 D, hepatocytes 7 days after partial hepatectomy C, 0' digestion-time control. Topological zones relative to the NM: D, distal; P, proximal; VC, very close; E, embedded within the NM. (-) Negative control (no template); (+) positive control (pure genomic DNA as template). The amplification patterns were consistently reproduced in separate experiments with samples from independent animals (n ≥ 3).