Figure 8.
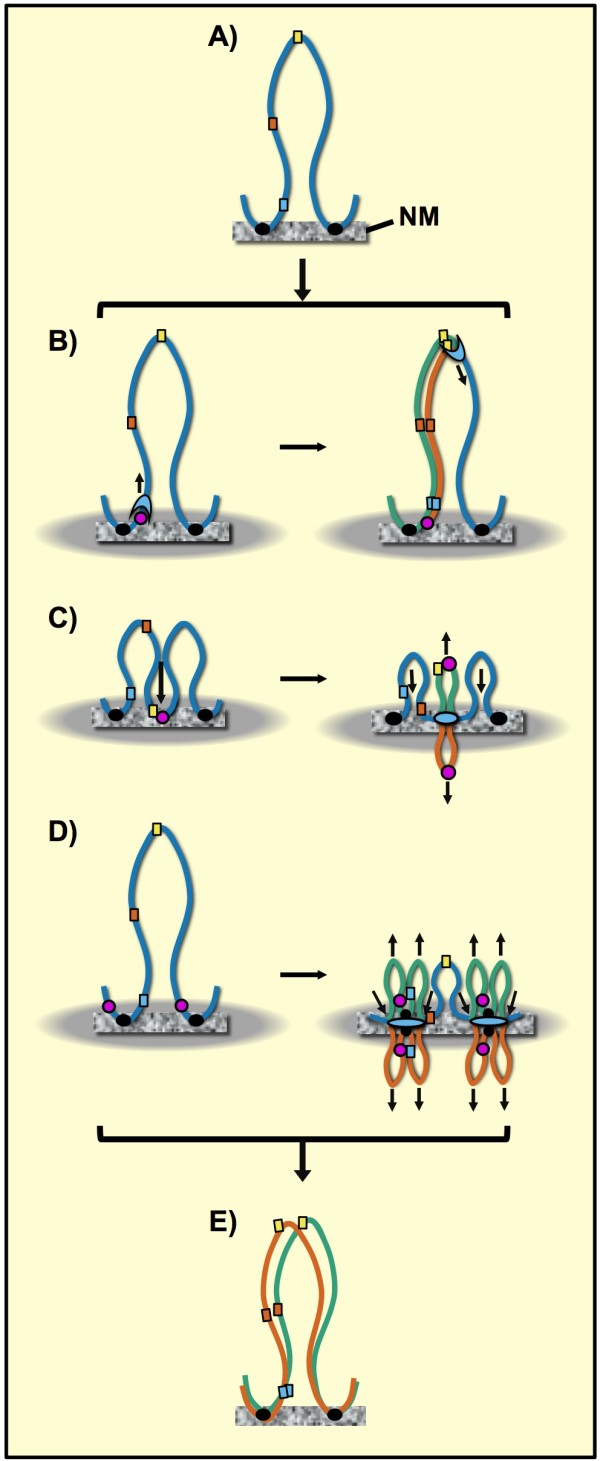
Possible mechanisms for replicating DNA loops in vivo. A) DNA loops are attached to the nuclear matrix (NM) by LARs sequences (black circles), the colored squares represent different sequences located along the loop DNA. B) Tracking model of DNA replication in which the replication complex (light blue) binds to a replication origin (purple circle) and moves along the template (blue string) while synthesizing the nascent daughter duplexes (orange and green strings). C) Reel-in model for DNA replication in which a replication origin (purple circle) originally at the tip of the DNA loop binds to the NM in late G1 reducing the actual loop size in half. The large grey oval represents the replication focus or factory. During the S phase the template (blue string) is pulled towards the replication complex on the NM that extrudes the newly replicated daughter duplexes (orange and green strings) that included the early-replicated origins. The LAR sequences (black circles) will be the last to be replicated. D) Reel-in model for DNA replication in which the origins are close to the LARs which are among the first loop DNA sequences to be duplicated. Replication begins close to the LARs but the newly-replicated LARs immediately attach to the NM as well as the newly-replicated origins that after initially firing gave origin to the actual replication forks (light blue ovals). Thus the growing daughter duplexes extruded from the replication complex on the NM are doubled in number, resulting in tiny but transient DNA loops that disappear once the loop DNA has been fully replicated, resulting in two full-sized daughter DNA loops (E).
