Figure 1.
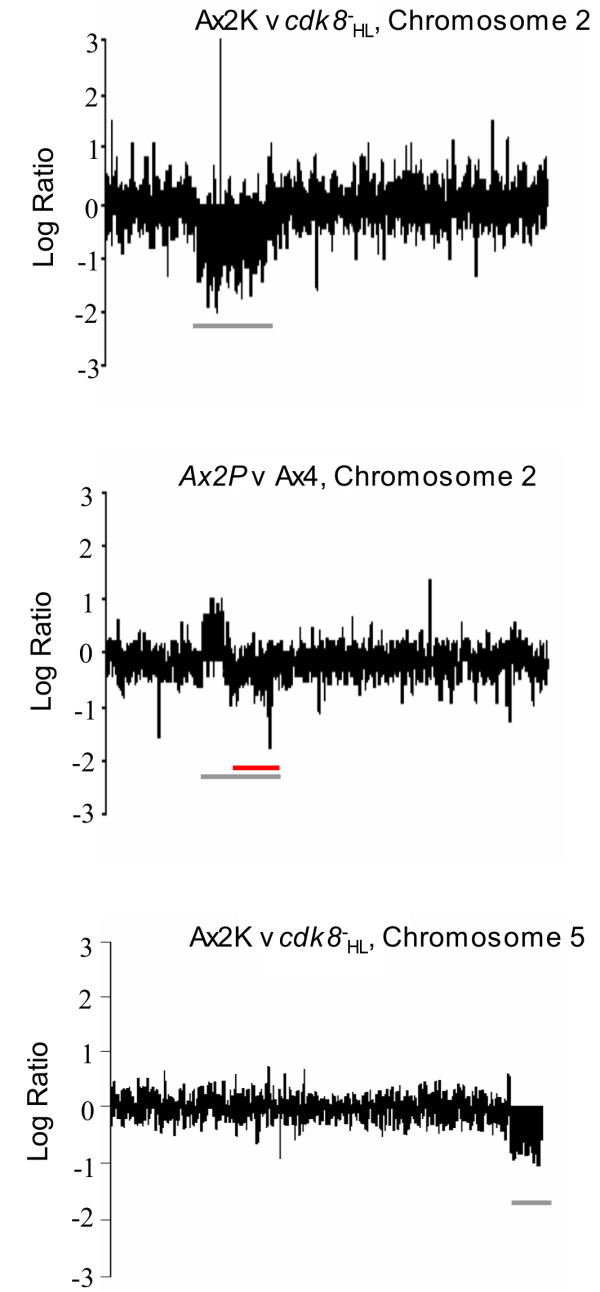
Microarray comparisons of genomic DNA from Ax2, Ax4 and cdk8-HL strains. The log(2)ratio of abundance of each gene in each comparison is plotted against the position of that gene on the chromosome. Ax2P is the parental strain used to generate cdk8-HL and Ax2K is the Ax2 strain with minimal genomic duplications [14]. Shown are comparison of genes on Chromosome 2 between Ax2K and cdk8-HL, and between Ax2P (the parent of cdk8-HL) and Ax4, and a comparison of genes on chromosome 5 for Ax2K and cdk8-HL. The approximate position of the putative cdk8-HL duplications relative to Ax2K are marked with grey horizontals bars. Logratios of approximately 1 or -1 indicate 2-fold changes in copy number (duplications); logratios around zero indicate equal copy number. The known Ax4 duplication is marked with a red horizontal bar; logratios in this region average approximately zero in this comparison show that both strains have the same copy number here. Ax2P has a longer duplication than that apparent in Ax4, though in the same region, as represented by the grey bar in the middle panel. The cdk8 gene is found on chromosome 1 and so its loss is not apparent on the comparisons shown here.
