Abstract
The 25 human bitter receptors and their respective genes (TAS2Rs) contain unusually high levels of allelic variation, which may influence response to bitter compounds in the food supply. Phenotypes based on the perceived bitterness of single bitter compounds were first linked to food preference over 50 years ago. The most studied phenotype is propylthiouracil bitterness, which is mediated primarily by the TAS2R38 gene and possibly others. In a laboratory-based study, we tested for associations between TAS2R variants and sensations, liking, or intake of bitter beverages among healthy adults who were primarily of European ancestry. A haploblock across TAS2R3, TAS2R4, and TAS2R5 explained some variability in the bitterness of espresso coffee. For grapefruit juice, variation at a TAS2R19 single nucleotide polymorphism (SNP) was associated with increased bitterness and decreased liking. An association between a TAS2R16 SNP and alcohol intake was identified, and the putative TAS2R38–alcohol relationship was confirmed, although these polymorphisms did not explain sensory or hedonic responses to sampled scotch whisky. In summary, TAS2R polymorphisms appear to influence the sensations, liking, or intake of common and nutritionally significant beverages. Studying perceptual and behavioral differences in vivo using real foods and beverages may potentially identify polymorphisms related to dietary behavior even in the absence of known ligands.
Keywords: alcohol drinking, alleles, bitter taste receptors, food choice, genetics
Introduction
The 25 human bitter receptors and their respective genes (TAS2Rs) contain unusually high levels of allelic variation, which may indicate local adaptation for the avoidance of plant toxins (Kim et al. 2005). Work on the relationship between bitter taste phenotypes and ingestive behavior dates back to the pioneering work of Fischer, Kaplan, and Glanville a half century ago (Fischer et al. 1961; Glanville and Kaplan 1965). Of these, the most studied is differential bitter response for phenylthiocarbamide (PTC) and propylthiouracil (PROP), which is mediated by the TAS2R38 gene (Kim et al. 2003) and possibly others (Hayes et al. 2008; Reed et al. 2010). For many years, work relating taste to food sensations focused on bitter phenotype as a monolithic entity that is high or low in an individual (e.g., Hall et al. 1975; Drewnowski et al. 1997), raising the obvious question of why individuals are particularly susceptible to the bitterness of some foods but not others. Recent advances in molecular taste genetics (e.g., Meyerhof et al. 2010) and psychophysics (e.g., Delwiche et al. 2001) show a complex and multifaceted response to bitter stimuli. Here, in a sample of adults tested in the laboratory, we provide preliminary evidence of novel associations between TAS2R genes beyond TAS2R38 and the bitterness of or ingestive behaviors toward several common bitter beverages—coffee, grapefruit juice, and alcohol—potentially explaining differential bitter response to foods across individuals. In the process, we also identify foods that may potentially contain candidate ligands for several orphan taste receptors.
The hT2R16 G protein–coupled receptor encoded by the TAS2R16 gene (Entrez GeneID: 50833) mediates the detection of salicin and other naturally occurring bitter compounds in the beta-glucopyranoside family (Bufe et al. 2002), and polymorphisms in this locus confer differential response in vitro via functional changes in the receptor (Soranzo et al. 2005). Prevalence of these alleles vary by geographic region, suggesting allelic variation may have arisen to as an evolutionary adaptation to the local plant environment (Soranzo et al. 2005). The less sensitive K172 ancestral allele is commonly found in African populations, whereas the more sensitive N172 variant is found in almost all individuals of European or Asian origin. In the contemporary environment, the less sensitive K172 allele appears to be a risk factor for alcohol intake (Wang et al. 2007) and dependence (Hinrichs et al. 2006). Because the K172 allele is rare among European Americans (minor-allele-frequency of 0.6%), we chose to test the association between 2 alternative TAS2R16 single nucleotide polymorphisms (SNPs) and alcohol intake among our sample of adults who are of predominantly European ancestry.
The Proline49–Alanine262–Valine296 (PAV) haplotype for TAS2R38 (Entrez GeneID: 5726) confers the ability to taste thiourea compounds like PROP and PTC at low concentrations; these compounds are part of a structurally distinct class from the bitter beta-glucopyranosides discussed above. The ancestral PAV allele protects against the ingestion of bitter thyroid toxins; the selective pressure for the Alanine–Valine–Isoleucine (AVI) nontaster variant is unknown (Wooding 2006), although it could allow the consumption of bitter phytonutrients that confer health benefits (Duffy 2007) as this polymorphism predicts vegetable bitterness (Sandell and Breslin 2006) and intake (Duffy et al. 2010). TAS2R38 variation also associates with alcohol intake (Duffy et al. 2004; Wang et al. 2007) but not dependence (Wang et al. 2007), and this relationship is presumably mediated through variable bitterness of alcoholic beverages (Lanier et al. 2005). We previously reported that AVI homozygotes consume more alcohol than heterozygotes, who in turn consume more alcohol than PAV homozygotes (Duffy et al. 2004). Subsequently, the Collaborative Studies on Genetics of Alcoholism (COGA) used family-based methods to link TAS2R38 polymorphisms with alcohol intake in African-Americans (Wang et al. 2007). Because the 2 studies differ significantly in terms of method and sample, here we have tested associations between TAS2R38 and TAS2R16 and alcohol intake, replicating our earlier finding (Duffy et al. 2004) and demonstrating a new TAS2R16 intake finding in a new cohort of healthy adults. Based on our finding that the endogenous sweetness and bitterness of whisky predicts alcohol intake (Lanier et al. 2005), we also tested whether TAS2R16 and TAS2R38 variation might explain perceived sensations from this alcoholic beverage.
PROP bitterness explains ∼13% of the variance in the bitterness of sampled espresso coffee (Lanier et al. 2005), yet caffeine bitterness is only minimally correlated with the bitterness of PROP (Delwiche et al. 2001; Keast and Roper 2007). Nonetheless, the perception of caffeine and PROP appears to share a common genetic factor (Hansen et al. 2006). Here, in an exploratory analysis, we test whether variants for TAS2R genes might explain variation in the perceived bitterness of sampled espresso coffee. Similarly, because PROP bitterness covaries with the bitterness (Lanier et al. 2005) and liking of grapefruit juice (Drewnowski et al. 1997; Lanier et al. 2005), we test whether common bitter receptor gene SNPs explain behavioral responses to grapefruit juice.
Materials and methods
Subjects and procedure
A convenience sample of healthy adults was recruited by poster and word of mouth from the University of Connecticut community, a rural college campus. Individuals who smoked more than 9 cigarettes per week were excluded because cigarette use may alter taste perception (Sato et al. 2002) and because cigarette use is confounded with alcohol consumption (Bottoni et al. 1997). The study sample of 96 adults was mostly of European ancestry (85%), female (76%) and of middle age (mean 40.9 years ± 12.2 standard deviation). They had 2 sessions in the taste laboratory and a separate visit for venipuncture for DNA analysis. The study sample was separate from that reported previously (Duffy et al. 2004). The University of Connecticut Institutional Review Board approved all procedures; subjects gave informed written consent and were paid for their participation.
Stimuli
Approximately 10 mL samples of unsweetened grapefruit juice (Veryfine Products), instant espresso (Café Bustelo; Rowland Coffee Roasters), and a blended scotch whisky (Dewar's White Label; John Dewar and Sons) were tasted. These samples were served at room temperature in plastic medicine cups in random order under normal lighting conditions. All subjects rinsed between samples with room temperature deionized (>15 MΩ) water. Throughout the testing sessions, subjects rated the intensity of a series of 1000 Hz tones as a cross-modal standard; they were presented in 12 dB steps (50–98 dB) as described elsewhere (Hayes and Duffy 2008).
Scaling
Intensity and hedonic scaling data were collected using the general Labeled Magnitude Scale (gLMS), a semantically labeled line scale (Green et al. 1996), anchored at the top (100) to “strongest imaginable sensation of any kind” (Bartoshuk et al. 2005). Intermediate verbal labels included “barely detectable” (1.4), “weak” (6), “moderate” (17), “strong” (35), and “very strong” (53). Subjects were oriented to the gLMS by rating the intensity of 16 remembered oral and nonoral sensations (thermal, light, and sound sensations) to encourage consistent use of the gLMS across multiple sensory domains. After scale orientation, subjects used the gLMS to rate liking/disliking and the sweetness, sourness, saltiness, and bitterness of sampled nonalcoholic and alcoholic beverages. Sourness and saltiness were not used in any analyses but were included in the tasting protocol to minimize halo-dumping response bias (Clark and Lawless 1994). When making hedonic ratings, subjects were told the bottom of the scale was neither like nor dislike, and the top was either the “strongest imaginable liking of any kind” or “strongest imaginable disliking of any kind.” Subjects verbally indicated whether they liked or disliked the sample. Gene effects on the sensations and liking of the bitter beverages were assessed via analysis of covariance (ANCOVA), with covariates of age, sex, and the average intensity of 86 dB tones.
Self-reported intake
Subjects reported the quantity and frequency of consuming beer, wine, and liquor, where a standard drink equaled 12, 5, and 1.5 oz, respectively on a validated semiquantitative food frequency survey (www.nutritionquest.com). We tested TAS2R associations with frequency of alcohol consumption (number of times per year) and total alcohol intake (frequency multiplied by servings per drinking episode), relying on frequency as the primary intake measure for 2 reasons: 1) taste gene effects may become attenuated as a drinking bout progresses and 2) if taste genotype is systematically related to beverage choice, using standard drinks might bias intake estimates given that mixed drinks are potentially more variable in alcohol content. Genetic effects on alcohol intake were assessed via ANCOVA, with age as a covariate.
DNA extraction and genetic analysis
DNA was extracted from whole blood following manufacturer's instructions (Gentra), with occasional modification for lysed samples. Purified DNA samples were stored at 4 °C in Tris 10 mM and EDTA acid 1 mM until analyzed. When necessary, DNA was amplified using the Illustra GenomiPhi V2 D amplification kit from GE Lifesciences. Genotypes were determined using vendor-supplied assays from Applied Biosystems (Supplement Table 1) with the plates read on an ABI Prism 7900 HT (Applied Biosystems). For TAS2R38, we assumed that individuals heterozygous at all 3 SNPs (e.g., genotypes CG TC CT) were common haplotype heterozygotes (e.g., PAV/AVI rather than AAV/PVI) as the probability of having 2 rare haplotypes is extremely low (Wooding et al. 2004). SNP frequencies in our sample were similar to values seen in reference data sets, and none of the SNPs showed significant deviation from Hardy–Weinberg equilibrium (Supplementary Table S2).
We present unadjusted and Bonferroni adjusted P values here for the coffee and grapefruit hypotheses as there is an active debate as to whether multiple comparison adjustments are appropriate for exploratory studies (see Bender and Lange 2001). Here, the P values were adjusted by multiplying observed P value by 10 (2 haplotypes plus 7 unrelated SNPs on chromosome 7 and 1 SNP on chromosome 12). For alcohol, tests related to TAS2R16 and TAS2R38 were separate a priori hypotheses to replicate prior work (Duffy et al. 2004; Wang et al. 2007), so no multiple comparisons adjustments were made. As population stratification may potentially cause false negatives and false positives in gene association studies (Hamer and Sirota 2000), we also analyzed our data excluding those individuals who were not of European ancestry; because none of the results were substantively different in this subset, results for the entire sample are presented here.
Results
Confirmation that TAS2R16 and TAS2R38 polymorphisms explained differences in alcohol intake
Over the last year, the self-reported frequency of consuming all types of alcoholic beverages was 102 ± 13 (mean ± standard error of the mean), with good variability in intake frequency across the sample (interquartile range [IQR]: 15–141). In terms of total beverage intake (quantity times frequency), the mean intake was 188 ± 28 standard drinks per year (IQR: 18–268). Out of total drinks reported by the sample, 45% were beer, 40% were wine, and 15% were spirits (neat or with mixers).
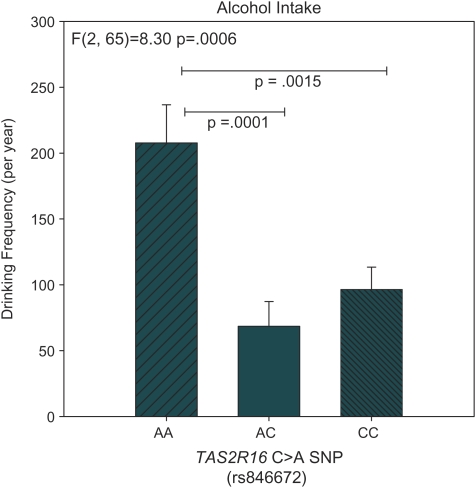
The TAS2R16 C>A (rs846672) SNP was significantly associated with frequency of consuming alcoholic beverages (F2,65 = 8.30, P = 0.0006); the minor allele homozygotes (AA carriers) consumed alcoholic beverages twice as frequently as the heterozygotes or major allele homozygotes, who did not differ in drinking frequency (Figure 1). A parallel analysis accounting for total beverage intake also indicated that the AA homozygotes drank more (F2,65 = 3.74 P = 0.029) than did the AG (P = 0.007) or GG (P = 0.039) carriers.
Figure 1.
Self-reported alcohol intake frequency in 68 adults. Effects of TAS2R16 SNPs on intake were assessed via ANCOVA, controlling for age (see text). Data represent means ± standard errors. Pairwise comparisons between groups (Fisher least significant difference) are indicated with horizontal lines. This figure appears in color in the online version of Chemical Senses.
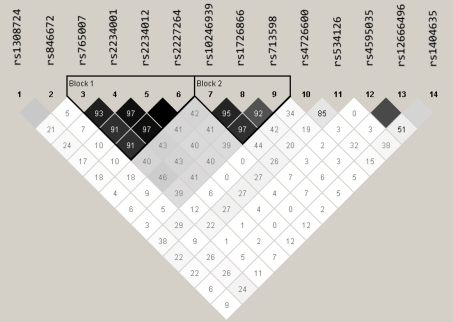
For the C>G SNP in TAS2R16 (rs1308724), there was a trend for CC homozygotes to consume alcohol less frequently (48 times/year) than did the heterozygotes (88 times/year), who consumed less frequently than did the GG homozygotes (122 times/year); however, this pattern was not significant for frequency (F2,64 = 2.05 P = 0.137) or total intake (F2,64 = 1.93 P = 0.15). Removing alcohol abstainers (those who reported no alcohol consumption over the year) improved the association between the C>G SNP and the 2 alcohol measures (P = 0.05 and 0.07). Because the 2 SNPs are not in linkage disequilibrium (LD; Figure 5), a larger sample is needed to rule out whether the C>G allelic variant (rs1308724) makes a separate contribution after accounting for the C>A (rs846672) effects.
Figure 5.
LD Plot for TAS2R SNPs on chromosome 7 showing R2 values generated via Haploview. Block 1 indicates a haploblock across TAS2R3, TAS2R4, and TAS2R5 (see text for details), and block 2 indicates the expected PAV/AVI haploblock for TAS2R38. This figure appears in color in the online version of Chemical Senses.
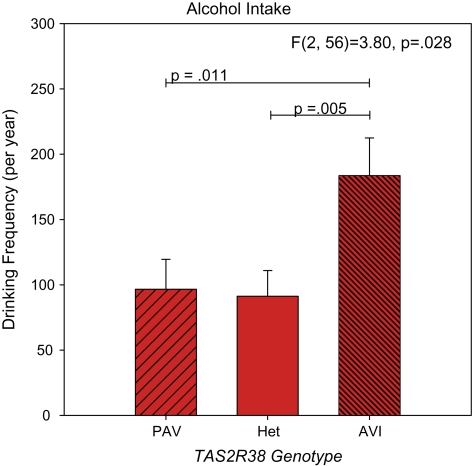
Here, we reaffirm the prior finding (Duffy et al. 2004) that AVI homozygotes drink alcohol more frequently than do PAV homozygotes (Figure 2). However, the heterozygotes here did not show an intermediate level of intake between the AVI and PAV homozygotes; it appears having one copy of the taster allele may be sufficient to depress intake (F2,56 = 3.80 P = 0.028). As above, using total beverage intake as the consumption variable showed consistent findings; AVI homozygotes drank more (F2,56 = 4.32 P = 0.018) than either heterozygotes (P = 0.004) or PAV homozygotes (P = 0.008).
Figure 2.
Self-reported alcohol intake frequency in 60 adults. Effects of TAS2R38 on intake were assessed via ANCOVA, controlling for age. Genotypes are PAV = PAV/PAV; Het = PAV/AVI; AVI = AVI/AVI; individuals with rare haplotypes were excluded. Data represent means ± standard errors. Pairwise comparisons between groups (Fisher least significant difference) are indicated with horizontal lines. These data are from a new sample of subjects, providing independent confirmation of our earlier report (Duffy et al. 2004). This figure appears in color in the online version of Chemical Senses.
As expected given the lack of LD between TAS2R16 and TAS2R38 (Figure 5), each contributed independently to the alcohol consumption measures in 2-way analysis of variance (p < 0.05). The small numbers in the subgroups do not warrant pairwise comparisons.
Polymorphisms in TAS2R16 or TAS2R38 failed to explain variability in taste sensations (sweetness, sourness, saltiness, and bitterness) elicited by the scotch whisky nor did they explain variation in liking (not shown).
SNPs in TAS2R3, TAS2R4, and TAS2R5 formed a haploblock that explained coffee bitterness
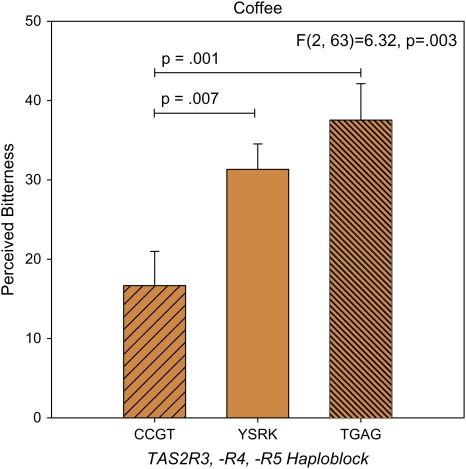
Four SNPs in TAS2R3, TAS2R4, and TAS2R5 each explained variation in the bitterness of coffee (not shown). Given this consistent pattern, and because these genes are located near each other on chromosome 7, we determined whether these SNPs formed a haplotype block (Figure 5) and found that 89% of our sample possessed one of 3 common haplotypes as shown below (consistent with unpublished data from Alarcon et al. 2008). Analyzed as a haplotype, allelic variation across TAS2R3, -R4, and -R5 explained variability in coffee bitterness (F2,63 = 6.32, P = 0.003, adjusted P = 0.03); individuals with 1 or 2 copies of the more responsive haplotype (TGAG) experienced twice as much bitterness compared with individuals homozygous for the less responsive haplotype (CCGT) (Figure 3). Notably however, the haploblock did not predict coffee liking (F2,63 = 1.31, P = 0.278).
Figure 3.
Bitterness intensity ratings of espresso coffee, collected using the gLMS, a psychophysical scale with ratio properties (i.e., a 40 is twice as great as a 20). Effects of the TAS2R3/R4/R5 haploblock were assessed via ANOVA. Genotypes are CCGT = CC,CC,GG,TT; YSRK = C or T, C or G, A or G, G or T; TGAG = TT, GG, AA, GG. Data represent means ± standard errors. Pairwise comparisons between groups (Fisher least significant difference) are indicated with horizontal lines. This figure appears in color in the online version of Chemical Senses.
| TAS2R3 | TAS2R4 | TAS2R5 | % Of sample | |
| rs765007 | rs2234001 | rs2234012 | rs2227264 | |
| C | C | G | T | 26% |
| Y | S | R | K | 41% |
| T | G | A | G | 22% |
| 5′UTR | Val96Leu | 5′UTR | Ser26Ile | |
| Y = C/T, S = C/G, R = A/G, K = G/T | ||||
TAS2R19 variation influenced grapefruit juice bitterness and liking
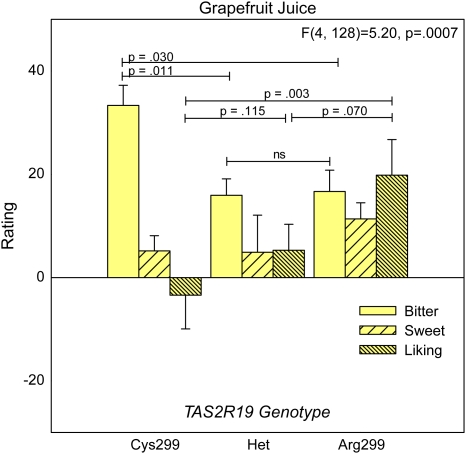
The rs10772420 A>G coding single nucleotide polymorphism (cSNP) in TAS2R19 results in the substitution of Arg for a Cys at amino acid position 299 (Cys299Arg), and this polymorphism appeared to influence grapefruit juice bitterness and thus sweetness and liking (F4,128 = 5.20, P = 0.0007, adjusted P = 0.007). Individuals who were homozygous for the Cys299 allele rated grapefruit juice twice as bitter as Arg299 homozygotes (P = 0.030) or heterozygotes (P = 0.011) (Figure 4). We found that the more sensitive Cys299 homozygotes (P = 0.003) and the heterozygotes (P = 0.070) liked the grapefruit juice less than the less sensitive Arg299 homozygotes. For the rs4595035 SNP in TAS2R60, similar patterns were observed for bitterness and liking (F4,128 = 2.49, P = 0.0469), but the effect was no longer significant after multiple comparisons adjustment (P = 0.47) suggesting this finding in particular requires independent confirmation in a larger sample to disentangle it from TAS2R19 effects.
Figure 4.
Bitterness, sweetness, and liking ratings of unsweetened white grapefruit juice. Effects of the TAS2R19 (rs10772420) SNP were assessed via ANOVA. Data represent means ± standard errors. Pairwise comparisons between groups (Fisher least significant difference) are indicated with horizontal lines. This figure appears in color in the online version of Chemical Senses.
Discussion
In sharp contrast to earlier work that viewed bitter response as a monolithic entity within in an individual (e.g., Hall et al. 1975; Drewnowski et al. 1997), present findings are wholly consistent with the modern view that bitter taste transduction occurs via a broad repertoire of T2R receptors, each of which are, for the most part, specific to particular classes of ligands (e.g., Bufe et al. 2002; Kuhn et al. 2004; Behrens et al. 2007; Pronin et al. 2007). To date, specific ligands have been identified for over half of the 25 hTAS2Rs (Ley 2008; Meyerhof et al. 2010), although only a handful are ecologically relevant ligands found in foods or beverages (e.g., Brockhoff et al. 2007; Intelmann et al. 2009). Here, we find preliminary evidence that polymorphisms in or near multiple TAS2R genes may influence the sensations, liking, or intake of common beverages that contain phytochemicals and other pharmacologically active ingredients linked to chronic diseases such as cardiovascular disease and cancer. Specifically, it appears that bitterness of alcoholic beverages, coffee, and grapefruit juice are mediated by multiple T2R receptor genes, and each of these genes appear to contain functional polymorphisms both within and across individuals, with the potential to influence intake.
Alcohol
Previously, Wang et al. (2007) reported an association between TAS2R38 haplotype and the maximum number of drinks in a 24-h period among Africans Americans but not European Americans. In contrast, present data generally confirm our prior finding that alcohol intake in European Americans is related to TAS2R38 genotype, although the question of whether these effects are additive (Duffy et al. 2004) or dominant (present data) are unresolved. The discrepancy in findings between our studies and Wang et al. could arise from using very different alcohol consumption measures (maximum drinks in 24 h vs. usual yearly consumption), the study sample (high-risk vs. reportedly healthy adults), or the analysis strategy (family based methods vs. explaining variance in intake among unrelated individuals).
Polymorphisms in TAS2R16 or TAS2R38 neither explained variability in taste sensations (sweetness, sourness, saltiness, and bitterness) elicited by blended scotch whisky nor liking for this alcoholic beverage. The genotype effects on alcohol consumption may be a generalized response to alcoholic beverages as a whole and not a sensory response to the particular specific alcoholic beverage tasted here. This suggests that any protective effect of TAS2R16 and TAS2R38 on alcohol intake may be unrelated to the bitterness of ethanol per se and could be due to other bitter ligands in other alcoholic beverages. Presumably, protective effects against intake generalize to all types of beverages via associative learning during early exposure. That is, heightened bitter response to ligands commonly found in some alcoholic beverages may depress the acquisition of preference for alcohol as a whole when an individual is first learning to consume alcohol. The most obvious candidate beverage would be beer, although this interpretation is complicated by the observation that hop-derived bitter tastants also fail to activate T2R16 and T2R38 expressing HEK cells in culture (Intelmann et al. 2009). Nonetheless, the present study is logically consistent with prior reports that alcohol behaviors in humans associate with certain bitter receptor alleles (e.g., Duffy et al. 2004; Wang et al. 2007) or bitter phenotypes (Intranuovo and Powers 1998; Lanier et al. 2005). This implies that taste receptor genes and bitter phenotypes may serve as useful biomarkers to study predisposition to chronic diseases associated with dietary behaviors (Sacerdote et al. 2007; Hayes 2010).
Coffee
The 4 SNPs across TAS2R3, -R4, and -R5 are located close to each other on chromosome 7, forming a haplotype block in our sample. The TGAG homozygotes perceived twice as much bitterness as CCGT homozygotes, with the heterozygotes falling in the middle. Within the CCGT/TGAG haploblock, 2 of the polymorphisms encode amino acid substitutions in TAS2R4 and -R5, whereas the other 2 occur in the 5′ untranslated region (5'UTR) of TAS2R3 and -R4. The C>G cSNP in TAS2R4 (rs2234001) results in Leu being substituted for Val at amino acid 96 (Val96Leu), whereas the T>G cSNP in TAS2R5 (rs2227264) results in Ile being substituted for Ser at amino acid 26 (Ser26Ile). The rs765007 SNP in TAS2R3 and the rs2234012 SNP in TAS2R5 are located in the 5'UTR, a region that typically contains sequences that regulate translation efficiency (or messenger RNA stability, although that is less likely to influence phenotype). Whether the variation in coffee bitterness seen for this haplotype results from functional alteration of T2R4 or T2R5 or influences on transcription that alter the expression of T2R3 or T2R4 is unclear from present data. As with all association data, these significant SNPs may simply be linked to other unmeasured polymorphisms that confer the functional effect. Regardless, the significant results indicate that there is a polymorphic property of that genomic region that affects the phenotype.
Notably, none of these polymorphisms were predictive of coffee liking. That variation in bitter receptor genes was unable to explain variation in liking is unsurprising: We have previously reported that coffee bitterness ratings do not predict liking (Lanier et al. 2005). Coffee flavor is a complex percept that depends on far more than bitterness (∼30 different odorants make critical contributions to its flavor (Czerny et al. 1999; Maeztu et al. 2001), and caffeine plays only a minor role in eliciting coffee bitterness (Frank et al. 2006). Moreover, liking for coffee flavor is altered by variable pharmacological effects of caffeine and prior experience (see Cines and Rozin 1982). As with alcohol, for some individuals, desirable pharmacological effects may override innate dislike of bitterness, regardless of intensity. Likewise, epistatic interactions may attenuate relationships between taste receptor polymorphisms and liking. For example, individuals may learn to dislike coffee flavor due to caffeine's anxiogenic properties in spite of carrying the less responsive haploblock found here. Because the anxiety elicited by caffeine varies with polymorphisms in the ADORA2A gene (Alsene et al. 2003; Childs et al. 2008), it would be prudent to stratify by the A2a allele in future work assessing the impact of the CCGT/TGAG haploblock on coffee liking and intake. Additionally, present findings may inform in vitro efforts to identify naturally occurring ligands for hT2R3 and hT2R5 as our findings suggest novel candidate ligands might be isolated from coffee.
Grapefruit juice
Individuals who carry the Arg299 allele for TAS2R19 found grapefruit juice to be less bitter. Reed et al. (2010) recently provided independent evidence that this polymorphism is functional in humans. Bitterness and sweetness suppress each other (Keast and Breslin 2003) and such mixture suppression occurs centrally not peripherally (Lawless 1979), suggesting that functional differences in bitter taste receptors may thus affect perceived sweetness, at least in beverages or foods that are perceptual mixtures. Consistent with this, we previously reported that individual differences in perceived sweetness and bitterness independently predict variation in grapefruit juice liking (Lanier et al. 2005). Here, we find that the more sensitive Cys299 carriers liked the grapefruit juice less than the less sensitive Arg299 homozygotes. Likewise, we found somewhat equivocal evidence of similar patterns of bitterness and liking for the TAS2R60 SNP. This is intriguing as the rs4595035 C>T substitution in TAS2R60 is a silent (synonymous) polymorphism. It was recently demonstrated that natural variation in “synonymous” codons may result in proteins with the same amino acid sequences that are functionally distinct (Kimchi-Sarfaty et al. 2007). Whether this is true for the highly polymorphic T2R family remains to be determined. Additionally, no ligand has been identified for TAS2R60 to date (Meyerhof et al. 2010), and TAS2R19 was recently implicated as a possible quinine receptor (Reed et al. 2010). Our findings suggest it may be worthwhile challenging cells expressing TAS2R19 (and possibly TAS2R60) receptors with taste-active compounds isolated from grapefruit juice to identify additional naturally occurring ligands. Notably, using dietary behavior as a rapid screen to identify potential ligands need not be limited to sampled stimuli. Previously, we analyzed self-reported grapefruit juice liking data obtained from a pencil and paper questionnaire and found similar effects of the Arg299Cys polymorphism in TAS2R19 (nee TAS2R48) (Duffy et al. 2009). This suggests that rapid hedonic surveys could help focus receptor-ligand discovery work toward ecologically relevant compounds found in specific foods. Additionally, as recently noted by Wooding et al. (2010), it seems likely that interactions between whole foods and bitter receptors involve multiple ligands simultaneously. Thus, using human behavioral response, in particular hedonics, complements functional assays that test single ligands in vitro by providing a more holistic view.
Conclusions
Collectively, we interpret these results to suggest that variation in perceived bitterness of alcoholic beverages, coffee, and grapefruit juice (e.g., regular foods and not pharmaceuticals like quinine or PROP) may be explained by common polymorphisms in TAS2R bitter receptor genes. Of course, as with any exploratory data on putatively functional polymorphisms, these associations need to be confirmed in an independent sample. Additionally, attempts to identify ecologically relevant receptor ligands may be informed by human behavioral data by narrowing the field of candidates to be surveyed. For example, present data suggest that the TAS2R19 and TAS2R60 gene products (the hT2R19 and hT2R60 receptors) should be interrogated in vitro with bitter taste active compounds found in grapefruit. In light of present findings, studies that attempt to link food choice, diet, and health to chemosensory variation need to take advantage of contemporary advances in molecular taste genetics and move beyond the historically narrow focus on thiourea (PROP and PTC) phenotypes and genetics, to include newly identified genetic variation. Additionally, more attention needs to be paid to other biological factors that influence dietary behaviors. For example, recent work indicates that fungiform papilla density (a proxy for receptor density) influences vegetable intake independently from TAS2R variation (Duffy et al. 2010). Given the wide diversity of TAS2Rs, single studies that fail to uncover links between specific bitter taste markers and liking for or intake of specific foods do not invalidate the broader hypothesis that chemosensory variation drives ingestive behaviors, with the potential to impact health and wellness. Cataloging these effects will require extensive effort to move forward.
Funding
Funding for this work was provided by United States Department of Agriculture Hatch Project CONS00827 funds and the National Institutes of Health DC008613 and AA007459. The authors wish to thank John McGeary and our anonymous reviewers for their insightful comments and feedback on this work.
Supplementary Material
References
- Alarcon S, Tharp A, Tharp C, Breslin PA. The effect of polymorphisms in 4 hTAS2R genes on PROP bitterness perception. Chem Senses. 2008;33:S43. [Google Scholar]
- Alsene K, Deckert J, Sand P, de Wit H. Association between A2a receptor gene polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology. 2003;28:1694–1702. doi: 10.1038/sj.npp.1300232. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Fast K, Snyder DJ. Differences in our sensory worlds: invalid comparisons with labeled scales. Curr Dir Psychol Sci. 2005;14:122–125. [Google Scholar]
- Behrens M, Foerster S, Staehler F, Raguse JD, Meyerhof W. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J Neurosci. 2007;27:12630–12640. doi: 10.1523/JNEUROSCI.1168-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender R, Lange S. Adjusting for multiple testing—when and how? J Clin Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- Bottoni A, Cannella C, Del Balzo V. Lifestyle and dietary differences in smokers and non-smokers from an Italian employee population. Public Health. 1997;111:161–164. doi: 10.1016/S0033-3506(97)00576-3. [DOI] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Massarotti A, Appendino G, Meyerhof W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J Agric Food Chem. 2007;55:6236–6243. doi: 10.1021/jf070503p. [DOI] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Childs E, Hohoff C, Deckert J, Xu K, Badner J, de Wit H. Association between ADORA2A and DRD2 polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology. 2008;33:2791–2800. doi: 10.1038/npp.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cines BM, Rozin P. Some aspects of the liking for hot coffee and coffee flavor. Appetite. 1982;3:23–34. doi: 10.1016/s0195-6663(82)80034-2. [DOI] [PubMed] [Google Scholar]
- Clark CC, Lawless HT. Limiting response alternatives in time-intensity scaling: an examination of the halo-dumping effect. Chem Senses. 1994;19:583–594. doi: 10.1093/chemse/19.6.583. [DOI] [PubMed] [Google Scholar]
- Czerny M, Mayer F, Grosch W. Sensory study on the character impact odorants of roasted arabica coffee. J Agric Food Chem. 1999;47:695–699. doi: 10.1021/jf980759i. [DOI] [PubMed] [Google Scholar]
- Delwiche JF, Buletic Z, Breslin PA. Covariation in individuals' sensitivities to bitter compounds: evidence supporting multiple receptor/transduction mechanisms. Percept Psychophys. 2001;63:761–776. doi: 10.3758/bf03194436. [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Shore AB. Taste responses to naringin, a flavonoid, and the acceptance of grapefruit juice are related to genetic sensitivity to 6-n-propylthiouracil. Am J Clin Nutr. 1997;66:391–397. doi: 10.1093/ajcn/66.2.391. [DOI] [PubMed] [Google Scholar]
- Duffy VB. Variation in oral sensation: implications for diet and health. Curr Opin Gastroenterol. 2007;23:171–177. doi: 10.1097/MOG.0b013e3280147d50. [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28:1629–1637. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Davidson AC, Kidd JK, Kidd KK, Bartoshuk LM. Vegetable intake in college-aged adults is explained by oral sensory phenotypes and TAS2R38 genotype. Chemosens Percept. 2010;3(3-4):137–148. doi: 10.1007/s12078-010-9079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Sullivan BS, Faghri P. Surveying food/beverage liking: a tool for epidemiological studies to connect chemosensation with health outcomes. Ann N Y Acad Sci. 2009;1170:558–568. doi: 10.1111/j.1749-6632.2009.04593.x. [DOI] [PubMed] [Google Scholar]
- Fischer R, Griffin F, England S, Garn SM. Taste thresholds and food dislikes. Nature. 1961;191:1328. doi: 10.1038/1911328a0. [DOI] [PubMed] [Google Scholar]
- Frank O, Zehentbauer G, Hofmann T. Bioresponse-guided decomposition of roast coffee beverage and identification of key bitter taste compounds. Eur Food Res Technol. 2006;222:492–508. [Google Scholar]
- Glanville EV, Kaplan AR. Food preference and sensitivity of taste for bitter compounds. Nature. 1965;205:851–853. [Google Scholar]
- Green BG, Dalton P, Cowart B, Shaffer G, Rankin K, Higgins J. Evaluating the 'Labeled Magnitude Scale' for measuring sensations of taste and smell. Chem Senses. 1996;21:323–334. doi: 10.1093/chemse/21.3.323. [DOI] [PubMed] [Google Scholar]
- Hall MJ, Bartoshuk LM, Cain WS, Stevens JC. PTC taste blindness and the taste of caffeine. Nature. 1975;253:442–443. doi: 10.1038/253442a0. [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L. Beware the chopsticks gene. Mol Psychiatry. 2000;5:11–13. doi: 10.1038/sj.mp.4000662. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCl, and caffeine. Chem Senses. 2006;31:403–413. doi: 10.1093/chemse/bjj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE. Response to “Lack of relation between bitter taste receptor TAS2R38 and BMI in adults”. Obesity (Silver Spring) 2010;18:433. doi: 10.1038/oby.2009.351. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JK, Duffy VB. Supertasting and PROP bitterness depends on more than the TAS2R38 Gene. Chem Senses. 2008;33:255–265. doi: 10.1093/chemse/bjm084. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Duffy VB. Oral sensory phenotype identifies level of sugar and fat required for maximal liking. Physiol Behav. 2008;95:77–87. doi: 10.1016/j.physbeh.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, et al. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78:103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intelmann D, Batram C, Kuhn C, Haseleu G, Meyerhof W, Hofmann T. Three TAS2R bitter taste receptors mediate the psychophysical responses to bitter compounds of hops (Humulus lupulus L.) and beer. Chemosens Percep. 2009;2:118–132. [Google Scholar]
- Intranuovo LR, Powers AS. The perceived bitterness of beer and 6-n-propylthiouracil (PROP) taste sensitivity. Ann N Y Acad Sci. 1998;855:813–815. doi: 10.1111/j.1749-6632.1998.tb10665.x. [DOI] [PubMed] [Google Scholar]
- Keast RS, Roper J. A complex relationship among chemical concentration, detection threshold, and suprathreshold intensity of bitter compounds. Chem Senses. 2007;32:245–253. doi: 10.1093/chemse/bjl052. [DOI] [PubMed] [Google Scholar]
- Keast SJR, Breslin PAS. An overview of binary taste-taste interactions. Food Qual Pref. 2003;14:111–124. [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 2005;26:199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier SA, Hayes JE, Duffy VB. Sweet and bitter tastes of alcoholic beverages mediate alcohol intake in of-age undergraduates. Physiol Behav. 2005;83:821–831. doi: 10.1016/j.physbeh.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Lawless HT. Evidence for neural inhibition in bittersweet taste mixtures. J Comp Physiol Psychol. 1979;93:538–547. doi: 10.1037/h0077582. [DOI] [PubMed] [Google Scholar]
- Ley JP. Masking bitter taste by molecules. Chemosens Percep. 2008;1:58–77. [Google Scholar]
- Maeztu L, Sanz C, Andueza S, De Pena MP, Bello J, Cid C. Characterization of espresso coffee aroma by static headspace GC-MS and sensory flavor profile. J Agric Food Chem. 2001;49:5437–5444. doi: 10.1021/jf0107959. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 2010;19(21):4278–4285. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdote C, Guarrera S, Smith GD, Grioni S, Krogh V, Masala G, Mattiello A, Palli D, Panico S, Tumino R, et al. Lactase persistence and bitter taste response: instrumental variables and mendelian randomization in epidemiologic studies of dietary factors and cancer risk. Am J Epidemiol. 2007;166:576–581. doi: 10.1093/aje/kwm113. [DOI] [PubMed] [Google Scholar]
- Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–R794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Sato K, Endo S, Tomita H. Sensitivity of three loci on the tongue and soft palate to four basic tastes in smokers and non-smokers. Acta Otolaryngol Suppl. 2002;546:74–82. doi: 10.1080/00016480260046445. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, et al. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcohol Clin Exp Res. 2007;31:209–215. doi: 10.1111/j.1530-0277.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- Wooding S. Phenylthiocarbamide: a 75-year adventure in genetics and natural selection. Genetics. 2006;172:2015–2023. doi: 10.1093/genetics/172.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Gunn H, Ramos P, Thalmann S, Xing C, Meyerhof W. Genetics and bitter taste responses to goitrin, a plant toxin found in vegetables. Chem Senses. 2010;35(8):685–692. doi: 10.1093/chemse/bjq061. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.