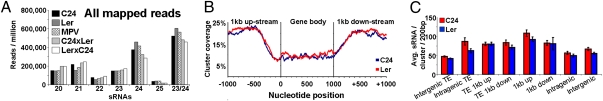
Fig. 2.
(A) sRNAs (20- to 25-nt) from total mapped population. An additional category (23/24-nt) was added, as these two classes likely represent a single class, generated at similar loci through identical biogenesis pathway, exerting similar downstream effects (16). (B and C) siRNA frequency and density over genes associated with siRNA clusters. (B) Probability of a given nucleotide being covered by a 24-nt siRNA cluster (gene body incorporates 5′ and 3′ UTR; position 0 indicates transcriptional start site). Only genes ≥1 kb were considered. All genes and respective coverage is scaled to 1 kb for parity. (C) Average siRNAs per cluster for a genomic feature (standardized to 200 bp, i.e., average cluster length). “TE 1kb up- or down-stream” is a TE situated within ±1 kb of a gene. Intergenic TEs are transposable elements residing at least ±1 kb from a gene. Error bars represent SEM.