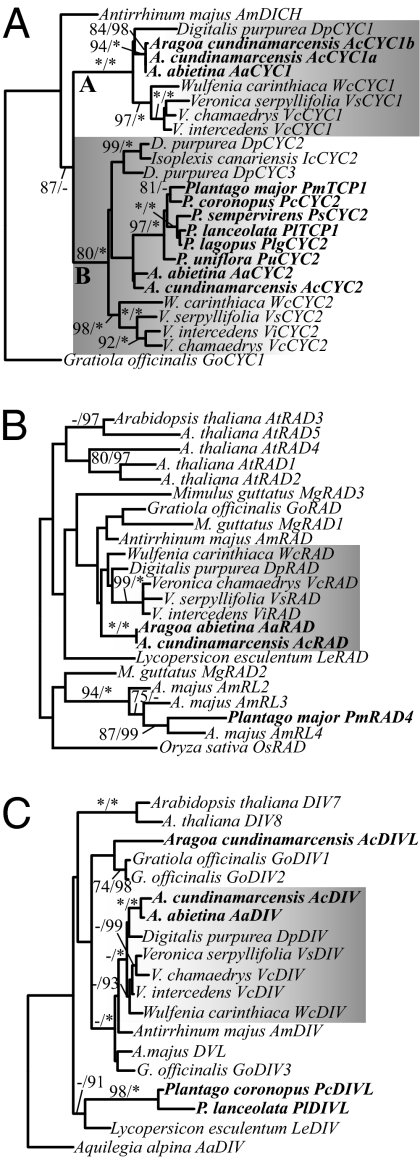
Fig. 2.
Maximum-likelihood phylograms showing best estimates of floral symmetry gene relationships. (A) CYC-like genes were duplicated following the divergence of Antirrhineae (represented by Antirrhinum majus) and Digitalideae (represented by Digitalis purpurea) + Veroniceae (represented by Wulfenia, Veronica, Aragoa, and Plantago), giving rise to two gene clades designated A and B (shaded). Plantago species have a single B-clade CYC-like gene (bold), but no genes were found within the CYC A clade, suggesting loss of this gene at the base of Plantago. (B) Plantago genomes have RAD-like paralogs (bold) but lack RAD orthologs. (C) Plantago genomes have DIV-like (DIVL) paralogs (bold) but lack DIV orthologs. Maximum-likelihood bootstrap values >70% (Left) and Bayesian posterior probabilities >90% (Right) are shown. Asterisks denote support values of 100%.