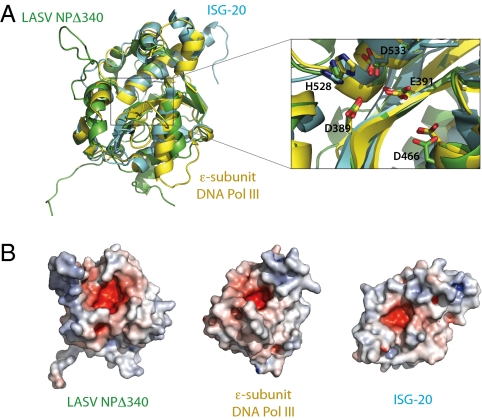
Fig. 2.
Superimposition of LASV NP and known DEDD exonucleases. (A) Structural comparison of NPΔ340 and two known DEDDh exonucleases. NPΔ340 is colored green, ISG-20 (PDB ID 1WLJ; ref. 27) is colored cyan, and the E. coli DNA pol IIIε (PDB ID 2GUI; ref. 28) is colored yellow. Inset shows a close-up view of the superimposed DEDDh residues of the active site. Numbered residues reflect those of LASV NP. (B) Electrostatic surface potential calculated with APBS (the Adaptive Poisson-Boltzmann Solver) software (49) shows that each exonuclease has an acidic active site and highlights the basic arm of LASV NP. Positive surface is colored blue; negative surface is colored red with limits ± 10 kT/e.