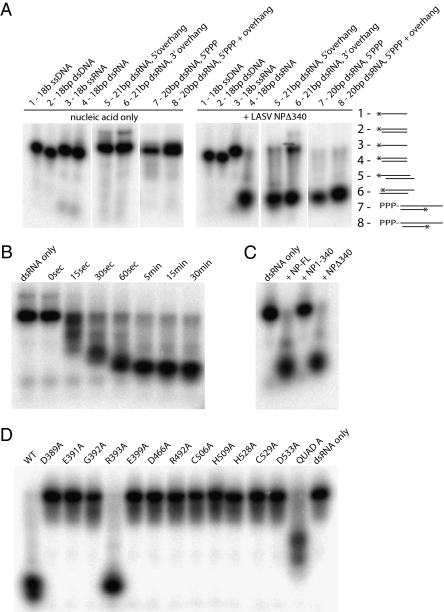
Fig. 3.
Ribonucleolytic activity of LASV NP. (A) Substrate specificity. NPΔ340 was incubated with different 32P-labeled substrates, and the reaction products were analyzed by denaturing PAGE and autoradiography. (Left) Migration patterns of each nucleic acid substrate alone (in the absence of NPΔ340 exonuclease). (Right) Migration of each substrate when incubated with NPΔ340 for 15 min. A schematic for each substrate is also shown, with the location of the 32P label indicated by an asterisk. Sequences for each substrate can be found in Table S2. (B) Time course of exonuclease activity. The 18-bp blunt-ended dsRNA is increasingly digested by wild-type NPΔ340 from 0 to 15 min. (C) Comparison of ribonucleolytic activity of the N terminus of NP (NP1–340) and full-length NP (NP-FL). The 18-bp blunt-ended dsRNA is digested by full-length NP and NPΔ340, whereas NP1–340 does not have exonuclease activity. (D) Effects of mutations to active site and proximal residues on ribonucleolytic activity. Wild-type and NPΔ340 point mutants were incubated with 18-bp blunt-ended dsRNA for 15 min, and products were analyzed by PAGE. QuadA designates a quadruple alanine mutation in residues K516K517K518R519. Note that all point mutations to residues in or near the exonuclease active site and the Zn coordination site, save R393, abrogate exonuclease activity, leaving the dsRNA undigested. The QuadA mutation to the basic arm partially diminishes exonuclease activity.