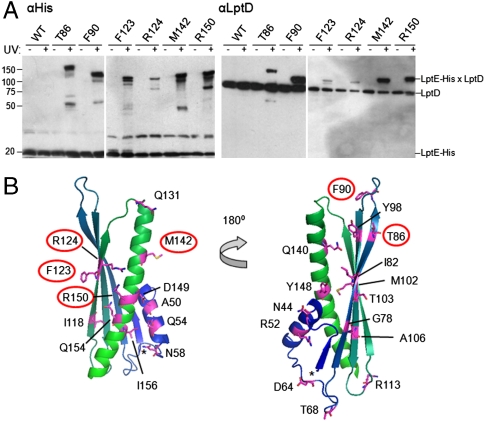
Fig. 2.
Specific residues of LptE interact directly with LptD in vivo. (A) Live E. coli expressing low levels of the indicated pBPA-substituted LptE–His protein were either left untreated or irradiated with UV light, followed by nickel affinity chromatography and Western blotting using an anti-His antibody or an anti-LptD polyclonal antiserum. (B) Mapping of the tested positions onto a structural model of E. coli LptE, generated on the basis of the three-dimensional structures of three LptE orthologs (RlpB from Shewanella oneidensis, Nitrosomonas europaea, and Neisseria meningitidis; PDB IDs 2R76, 2JXP, and 3BF2, respectively) using HHPred and MODELLER (40). Residues involved in LptD/E interactions, as identified in (A), are circled. The asterisk denotes the N-terminus of the model structure, corresponding to residue I36 of the full-length E. coli LptE. The N-terminal lipidation site is on C19 in this numbering scheme.