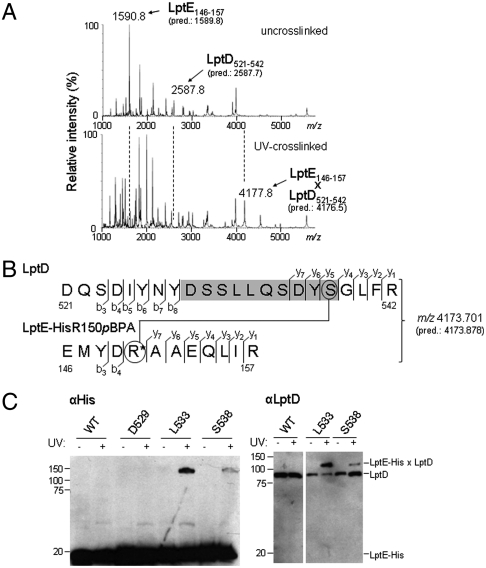
Fig. 3.
LptE interacts with a predicted extracellular loop of the LptD β-barrel. (A) LptD/E complexes in which LptE residue R150 was substituted with pBPA (LptE-HisR150pBPA) were overexpressed, purified, and UV-photocrosslinked in vitro (see Fig. S2A). Bands corresponding either to free LptD and LptE protein from uncrosslinked complexes (Top) or to the UV-crosslinked LptD/E complex (Bottom) were excised from an SDS-PAGE gel and subjected to in-gel trypsin digestion, followed by low-resolution MALDI-MS analysis of peptide products. The observed and predicted (pred.) average [M + H]+ masses of the indicated LptD and LptE peptides are shown. (B) High-resolution MS/MS fragmentation analysis of the crosslinked adduct identified in part A. The observed and predicted (pred.) exact [M + H]+ masses of this adduct are shown. Detected fragment ions included those corresponding to the eight N-terminal and four C-terminal residues of the LptD peptide spanning residues 521–542 (denoted b3–b8 and y1–y4, respectively, for the LptD sequence), as well as the four N-terminal and seven C-terminal residues of the pBPA-containing LptE peptide spanning residues 146–157 (denoted b3–b4 and y1–y7, respectively, for the LptE sequence; R* denotes the pBPA-substituted residue). Three additional C-terminal LptD fragments (y5–y7) had masses indicating the presence of a covalent modification on S538 (circled, Top) with molecular weight equal to that of the entire LptE peptide. No unmodified C-terminal LptD fragments of more than four residues were detected. (See Fig. S3 and Table S1.) (C) The indicated residues of LptD were replaced with pBPA in a chromosomal ΔlptD background and in the presence of plasmid-encoded LptE–His. UV-photocrosslinking, nickel affinity chromatography, and Western blotting were performed as in Fig. 2.