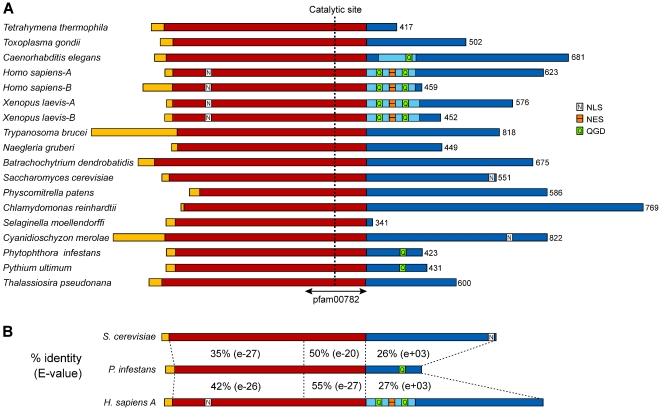
Figure 6. Structures of Cdc14 proteins.
(A) Proteins from the species in Table 1. The sequences are taken from their respective genome databases, except for the Naegleria, Selaginella, Trypanosoma, and Thalassiosira proteins which are based on manually curated gene models. The predicted proteins range from 341 to 822-aa as marked to the right of each model. Following a N-terminal region that shows little similarity between the proteins (yellow), each protein contains a fairly conserved stretch of about 300 aa (red). The latter includes the phosphatase domain which is marked as pfam00782, with the catalytic residue indicated. The C-terminal portions of the proteins (blue) show little conservation except for a roughly 85 aa region that is fairly conserved between C. elegans, human, and X. laevis (light blue). This includes the nuclear exit sequence (NES) and one or two QGD repeats. Nuclear localization signals (NLS) are also marked as detected by PSORTII; these include an experimentally validated NLS near the C-terminus of the S. cerevisiae protein [45], NLSs in the N-terminal regions of the human and X. laevis proteins which appear to have functions based on mutagenesis studies [20], [46], and a NLS predicted in the C-terminal region of the C. merolae protein. (B) Similarity between Cdc14 of P. infestans, S. cerevisiae, and human Cdc14A. The program SSEARCH was used to calculate the percent amino acid identity in the region upstream, upstream, and C-terminal to the pfam00782 phosphatase domain. E-values for each match are also provided, which indicate that the similarity at the C-terminus is insignificant.