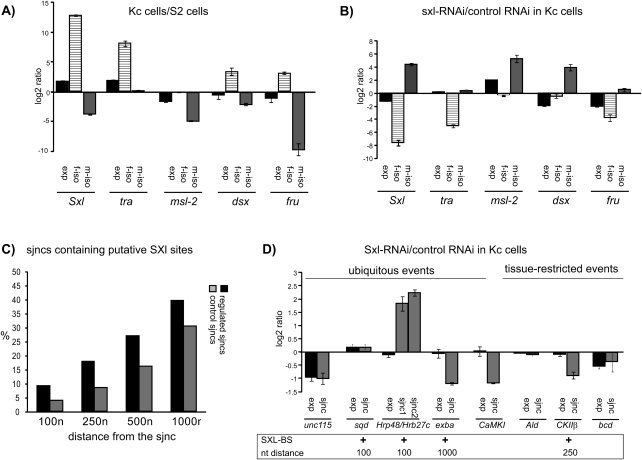
FIGURE 3.
Regulation of sex-specific splicing by SXL. (A) Alternative splicing pattern of the classical sex regulators (Sex-lethal [Sxl], transformer [tra], male specific lethal-2 [msl-2], doublesex [dsx] and fruitless [fru]) in female Kc and male S2 cells quantified by RT-PCR and represented as a ratio between the levels of male (m-iso) and female (f-iso) isoforms in the two cell lines. (exp) Gene expression values. (B) Reversal of sex-specific splicing patterns of sex determination genes in Kc cells upon Sex-lethal knockdown. Analysis of gene expression as well as of female- or male-specific isoforms was carried out as in A and represented as a ratio between RNAs isolated from cells in which Sex-lethal RNA was knocked down by RNAi and control RNAi. (C) Bioinformatic analysis of potential SXL-binding sites in 700 sex-regulated (black bars) and 700 non-sex-regulated sjncs (gray bars) in a window of 100, 250, 500, and 1000 nt adjacent to the splice site (see Materials and Methods for search criteria). (D) Quantitative-RT-PCR analysis of the effect of Sxl knockdown in Kc cells on alternative splicing of genes predicted by the microarray analysis. Analyses were carried out as in B and results of sex-specific differences are represented as in previous figures for eight genes: Unc-115, hrp40/sqd, Hrp48/Hrb27c, exba, and CaMKI from the group of genes showing ubiquitous sex-specific regulation and Ald, CKIIβ, and bcd from the tissue-restricted category. The presence of computationally predicted SXL binding sites is indicated below the graph, including the distance window in nucleotides (nt) from the splice site within which the SXL binding site is located.