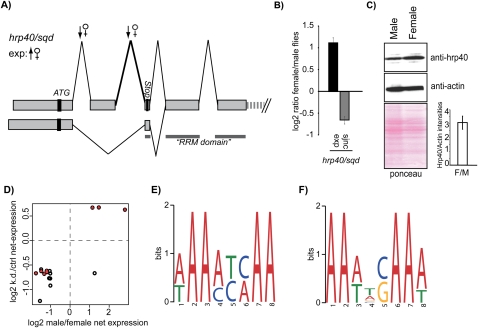
FIGURE 4.
A function for hrp40/squid in sex-specific splicing. (A) Schematic representation of splicing patterns of the hrp40/squid gene and splice junctions differentially regulated in female flies. ATG indicates the translational start site. Inclusion of exon 2 leads to an mRNA containing a premature stop codon (the pattern that is down-regulated in female flies), while the skipping of exon 2 produces the mRNA encoding full-length hrp40/squid protein. The gray line indicates exonic regions encoding the RNA binding domain (RRM). Only the sizes of exons are at scale. (B) Validation by quantitative RT-PCR of sex-specific differences in expression and alternative splicing of the hrp40/squid gene predicted by the microarray. Expression (exp) was quantified by amplification of a constitutive exon; sjnc indicates real-time PCR quantification using as one of the primers a splice-junction oligonucleotide corresponding to the junction indicated by the thicker line in A. (C) Western blot comparing levels of Hrp40/Squid protein in male and female flies (top) and loading controls (β-actin) (middle) and ponceau staining (bottom). Quantification of the relative levels of Hrp40/Squid vs. actin in female vs. male flies (F/M) for three independent experiments is shown in the lower-right panel. (D) Comparison of splice junction changes between hrp40/sqd knockdown vs. ctrl and male vs. female. Circles represent the intersection between the fold changes observed for a particular splice junction in the two experimental setups. Red triangles indicate splice junctions annotated as the alternative. Clustering at the lower-left and upper-right quadrants indicates correlation in the direction of splicing changes. (E) Top-enriched motif identified in exonic regions proximal to sex-specifically regulated sjncs. (F) Top-enriched motif identified in all regions proximal to sjncs differentially regulated upon hrp40/squid knockdown (Blanchette et al. 2009).