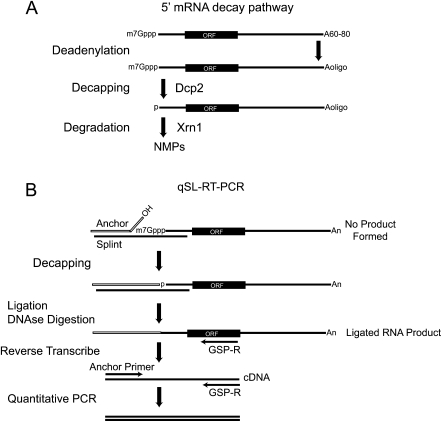
FIGURE 1.
The 5′ mRNA decay pathway. (A) mRNAs possess a 5′ 7-methyl guanosine cap (7mGppp), open reading frame (ORF) and a 3′ poly-adenosine tail, averaging 60–80 nt in yeast. mRNA decay via the 5′ pathway initiates with deadenylation of the poly-Adenosine tail to a short oligo-adenylated form (Aoligo). Dcp2 decapping enzyme subsequently removes the cap. The decapped mRNA, with a 5′ monophosphate, is destroyed by Xrn1, producing monophosphorylated nucleotides (NMP). (B) Schematic of the quantitative splinted-ligation reverse transcriptase polymerase chain reaction assay (qSL-RT-PCR). First, Anchor RNA and complementary DNA Splint oligonucleotides are annealed to the 5′ end of the target mRNA. mRNAs containing a 5′ cap cannot be ligated, as the cap prevents ligation. Decapped RNAs have a 5′ monophosphate that is ligated to the Anchor RNA 3′ hydroxyl by T4 DNA ligase. After ligation, the splint is destroyed by DNase I. The ligated RNA is converted to cDNA by reverse transcription with a reverse gene-specific primer (GSP-R). The resulting cDNA is then detected by quantitative PCR using GSP-R and a forward primer that anneals to the anchor (Anchor Primer). An internal control qPCR is performed on the same cDNA samples using gene-specific primers that amplify within the coding sequence of the mRNA.