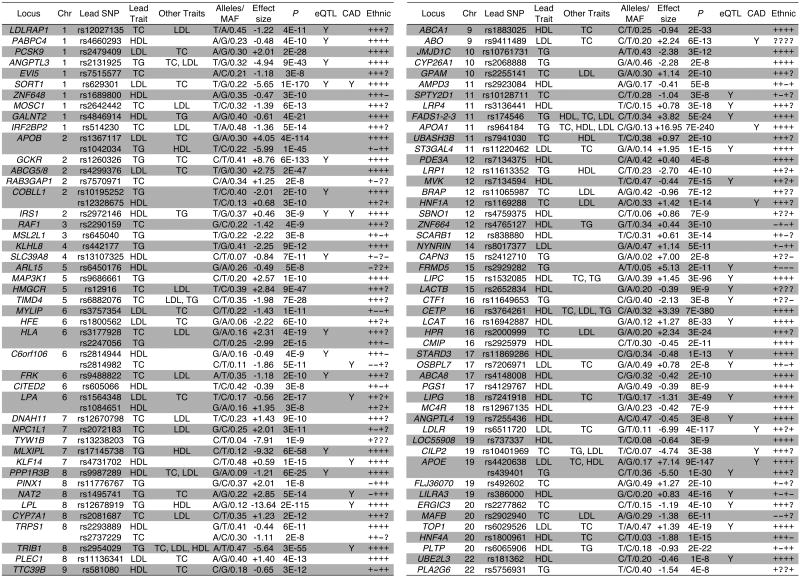
Figure 1. Meta-analysis of plasma lipid concentrations in >100,000 individuals of European descent.
The gene name listed in “Locus” column is either a plausible biological candidate gene in the locus or the nearest annotated gene to the lead SNP. Listed in “Lead Trait” column is the lipid trait with best P-value among all four traits. Listed in “Other Traits” are additional lipid traits with P < 5 × 10-8. Listed in “Alleles/MAF” column are: major allele, minor allele, and minor allele frequency (MAF) within the combined cohorts included in this meta-analysis (alleles designated with respect to the “+” strand; Supplementary Table 2). Numbers in “Effect Size” column are in mg/dL for the lead trait, modeled as an additive effect of the minor allele. P-values are listed for the lead traits. In the “eQTL” column, “Y” indicates that lead SNP has an eQTL with at least one gene within 500 kb with P < 5 × 10-8 in at least one the three tissues tested (liver, omental fat, subcutaneous fat). In the “CAD” column, “Y” indicates that the lead SNP meets the pre-specified statistical significance threshold of P < 0.001 for association with CAD and being concordant between the direction of lipid effect and the change in CAD risk. In the “Ethnic” column, “+” indicates concordant effect on lead trait of the variant between the primary meta-analysis cohort and the European or non-European group, “−” indicates discordant effect on lead trait, and “?” indicates data not available for the group; in order, the ethnic groups are European, East Asian, South Asian, and African American (Supplementary Table 11).