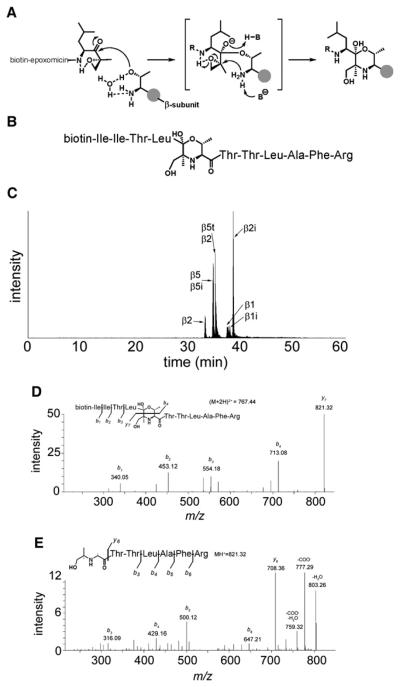
Figure 3. Active-Site Peptide Identification and Determination of Proteasome β Subunits by Affinity Purification, Tryptic Digest, and LC-MS Analysis.
(A) Reaction mechanism of biotin-epoxomicin 3 with the catalytically active N-terminal Thr residue of active proteasome β subunits. The morpholino ring formation results in a covalent and irreversible binding.
(B) Schematic representation of the biotin-epoxomicin modified, N-terminal active-site tryptic peptide of β5t. Amino acid residues are represented in a three-letter code.
(C) LC-MS elution profile of the six unique biotinylated tryptic peptides derived from the active sites. Notice that β5 and β5i active-site peptides are identical (see also Table S3).
(D) LC-MS2 determination of the β5t active-site fragmentation pattern. The parent ion [m/z (M+2H)2+ = 767.44] was fragmented. The b1, b2, b3, and b4 ions are signature ions of the biotin-epoxomicin N-terminal part. The abundant y7 ion containing the β5t active-site peptide sequence was selected for further (MS3) fragmentation (see E).
(E) LC-MS3 determination of the y7 ion (MH+ = 821.32) revealing the β5t active-site peptide amino acid sequence.