FIGURE 8.
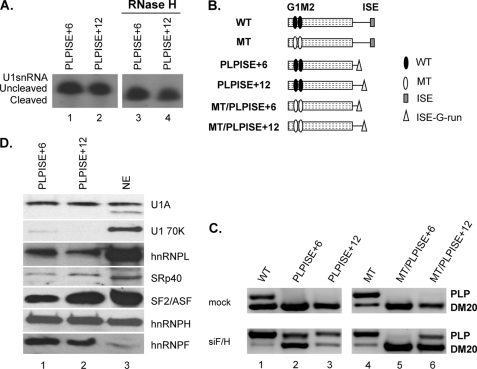
The function of the ISE is dependent on context and distance from the PLP 5′ splice site. A, representative primer extension analysis of U1snRNA bound to the PLP RNAs. Untreated (lanes 1 and 2) and RNase H-treated (lanes 3 and 4) HeLa nuclear extracts were incubated with the biotinylated RNA templates indicated (sequences in Table 1). The RNA was extracted from the streptavidin bead precipitates and assayed by reverse transcription with a primer complementary to loop 2 of the U1sn RNA (n = 2). The extension products from cleaved and uncleaved U1snRNA are shown. B, schematic representation of all constructs (see also Fig. 1). C, representative RT-PCR analysis of PLP and DM20 products amplified in RNA isolated from Oli-neu cells transfected with WT, PLPISE+6, PLPISE+12, MT, MT/PLPISE+6, and MT/PLPISE+12 and treated with siF/H (n = 3) (30 PCR cycles). Mock are cells transfected with the same plasmids and treated with scrambled siRNA (30 PCR cycles). D, representative Western blot analysis of RNA affinity precipitates of PLPISE+6 and PLPISE+12 (lanes 1 and 2, respectively) (sequences in Table 1) incubated with untreated HeLa nuclear extract (n = 2). Lane 3 is the HeLa nuclear extract. The precipitates were separated by SDS-PAGE, blotted, and probed with antibodies to U1A, U170K, SF2/ASF, SRp40, hnRNPL, hnRNPH, and hnRNPF.