Abstract
The flagellated protozoan Giardia duodenalis is a parasite of the upper part of the small intestine of mammals, including humans, and an interesting biological model. Giardia harbors a single 14-3-3 isoform, a multifunctional protein family, that is modified at the C terminus by polyglycylation, an unusual post-translational modification consisting of the covalent addition of one or multiple glycines on the γ-carboxyl groups of specific glutamic acids. Polyglycylation affects the intracellular localization of g14-3-3, as the shortening of the polyglycine chain is correlated with a partial relocalization of 14-3-3 inside the nuclei during encystation. In this work we demonstrate that the gTTLL3, a member of the tubulin tyrosine ligase-like family, is the enzyme responsible for the 14-3-3 polyglycylation. We also identify two metallopeptidases of the M20 family, here termed gDIP1 (giardial dipeptidase 1) and gDIP2, as enzymes able to shorten the g14-3-3 polyglycine tail both in vivo and in vitro. Finally, we show that the ectopic expression of gDIP2 alters the g14-3-3 localization and strongly hampers the cyst formation. In conclusion, we have identified a polyglycylase and two deglycylases that act in concert to modulate the stage-dependent glycylation status of the multifunctional regulatory g14-3-3 protein in G. duodenalis.
Keywords: Differentiation, Enzyme Purification, Eukaryote, Parasite, Post-translational Modification, Protozoan, 14-3-3 Protein, Giardia duodenalis, Encystation, Polyglycylation
Introduction
The flagellated and binucleated protozoan Giardia duodenalis (synonymous of Giardia lamblia and Giardia intestinalis) is an extracellular parasite of the upper part of the small intestine of mammals, including humans, where it causes giardiasis, an acute enteritis (1). Besides its relevance for human and animal health, G. duodenalis is also a fascinating and simple eukaryotic organism with a minimalistic genomic and cellular organization that arouses a great interest as a biological model (2). In this perspective we have previously characterized the single giardial 14-3-3 (g14-3-3) isoform, a member of a small dimeric protein family ubiquitously conserved in eukaryotes (3, 4). The 14-3-3s are able to bind a wide range of proteins containing consensus binding motifs usually phosphorylated on serine or threonine, thus, regulating multiple cellular processes, i.e. the metabolism, cell cycle progression, signal transduction pathways, cell growth, and differentiation (5). We demonstrated that the g14-3-3 is modified in a peculiar fashion by the phosphorylation of Thr-214 and the polyglycylation of Glu-246 (3, 4). The glycylation, first discovered at the C-terminal domain of α- and β-tubulin, is a post-translational modification consisting of the covalent addition of one or multiple glycines to the γ-carboxyl groups of specific glutamic acids of target proteins (6, 7). Recently, polyglycylation has been also reported for several proteins, including the mammalian nucleosome assembly proteins (8–10). Whereas the phosphorylation of g14-3-3 is a constitutive post-translational modification, the polyglycylation of the protein is regulated during the G. duodenalis life cycle with a remarkable reduction in the length of the polyglycine chain during the early phase of the encystation process (3, 4). Polyglycylation has been related to the intracellular localization of g14-3-3, as the shortening of the polyglycine chain is correlated with a partial relocalization of the protein inside the nuclei. In fact, in G. duodenalis parasites expressing the g14-3-3 mutant E246A, which cannot be polyglycylated, the protein localizes in the nuclei throughout the parasite life cycle, resulting in a faster differentiation of the trophozoite into the cyst stage once the process has been induced (3, 4).
Furthermore, the enzymes that catalyze the glycylation on tubulin and on other substrate proteins have been identified as members of the tubulin tyrosine ligase-like (TTLL)2 family, which also includes other amino acids ligases such as the tubulin tyrosine ligase (TTL) (11) and polyglutamylases, which add glutamic acid instead of glycines (12–14). Glycylases can be classified as: primases, which add the first glycine, like TTLL3s of vertebrates and Tetrahymena thermophila and the mammalian TTLL8 (9, 10, 15); elongases, which are only able to elongate the polyglycine chain, like mammalian TTLL10, with the exception of the non-functional human TTLL10 (9, 10); bifunctional initiating/elongating enzymes, like Drosophila melanogaster dmTTLL3A and dmTTLL3B (10). However, the existence of enzymes responsible for the removal of glycines has only been indirectly demonstrated, but information about their identity is still missing (16, 17).
In this work we demonstrate that the giardial TTLL3 (gTTLL3), a member of the TTLL family, is the enzyme responsible for the 14-3-3 polyglycylation. We also identify two metallopeptidases of the M20 family, here termed gDIP1 and gDIP2, as enzymes able to shorten the g14-3-3 polyglycine tail both in vivo and in vitro. Finally, we show that the ectopic expression of gDIP2 alters the g14-3-3 localization and strongly hampers the cyst formation.
EXPERIMENTAL PROCEDURES
Parasite Cultivation and Transfection
Trophozoites of the G. duodenalis strain WB-C6 were axenically grown for 72 h at 37 °C in the TYI-S-33 medium supplemented with 10% bovine serum and bovine bile at pH 7.0. Parasites were harvested by chilling culture tubes on ice for 30 min to detach adhering cells and centrifugation at 800 × g. Transgenic G. duodenalis lines were generated by electroporation in the presence of 15 μg of plasmid DNA and selection in the presence of 100 μm puromycin (Invivogen, Toulouse, France). Transgenic lines were maintained under constant selection with 100 μm puromycin. Encystation was induced as previously described (3).
Nucleic Acid Isolation
Genomic DNA of G. duodenalis WBC6 clone was isolated from 109 trophozoites as previously described (3). Total RNA was extracted from 107 trophozoites, or encysting parasites, using the RNAeasy mini kit (Qiagen, Hilden, Germany) following the manufacturer‘s instructions. Plasmid DNA was isolated from bacteria using the QIAPrep kit (Qiagen).
Vector Construction
Escherichia coli JM109 competent cells were used for vector manipulation and propagation. The full-length coding sequences of the seven giardial genes containing the TTLL motive (Prosite PDOC51221) and here arbitrarily termed gttll1 (GL50803_95661), gttll2 (GL50803_10382), gttll3 (GL50803_8456), gttll4 (GL50803_8592), gttll5 (GL50803_9272), gttll6 (GL50803_10801), and gttll7 (GL50803_14498) and the full-length coding sequences of the metallopeptidase-encoding genes gdip1 (GL50803_15832) and gdip2 (GL50803_8407) have been PCR-amplified from the genomic DNA of G. duodenalis WB-C6 clone using the primers listed in supplemental Table 1. PCR reactions were performed in a final volume of 50 μl using 5 μl of 10× buffer (Stratagene, La Jolla, CA), 50 μm dNTPs (Stratagene), 20 pmol of each primer, and 2.5 units of Pfu Ultra High Fidelity (Stratagene). Reactions were performed on a T-Personal Thermocycler (Biometra Corp., Göttingen, Germany). Amplification conditions were 1 cycle at 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min, and 1 cycle at 72 °C for 7 min. For the in vivo expression in G. duodenalis parasites, the full-length PCR fragments of the gTTLL were digested with NotI and BamHI or with BclI and cloned in the BamHI/PspOMI-digested pTUB-FLAGpac vector (see the supplemental Experimental Procedures), whereas the BclI/NotI-digested PCR fragments of gDIP1 gene and the BamHI/NotI-digested PCR fragments of gDIP2 gene were cloned in the BamHI/PspOMI-digested pTUB-FLAG-HApac vector (see the supplemental Experimental Procedures).
RNA Reverse Transcription and Real-time Quantitative PCR
For the cDNA synthesis, 2 μg of total RNA extracted either from trophozoites or encysting parasites at different time points as previously described were pretreated with 1 unit of DNase (Promega Corp., Madison, WI) for 30 min at 37 °C according to the manufacturer. Then the RNA was incubated with 4 units of Omniscript reverse transcriptase (Qiagen), 1 μm dT18 primer, 0.5 μm dNTP, 10× reaction buffer, and 10 units of RNase inhibitor (Promega) in a final volume of 30 μl for 2 h at 37 °C. Real-time PCR was performed in a LightCycler 480 (Roche Diagnostics). Amplification was performed in a final volume of 20 μl containing 6–10 ng of cDNA, 0.3 μm each of sense and antisense primers, and 10 μl of 2× LightCycler 480 SYBR Green Master (Roche Diagnostics). The selected genes were amplified using the primers listed in supplemental Table 2. The amplification program was 1 cycle of 5 min at 95 °C and 45 cycles of 12 s at 95 °C, 12 s at 59 °C, and 6 s at 72°. The final mRNA levels of the studied genes were normalized to gap1 gene expression using the 2−ΔΔCt relative quantification method (Roche Diagnostics). Graphed values were estimated from five independent experiments each repeated in triplicate.
Expression and Purification of the Recombinant Proteins
E. coli-transformed cells were grown in SOB medium (2% bacto tryptone, 0.5% yeast extract, 0.05% NaCl, 5 mm KCl, 10 mm MgCl2, pH 7.2) at A600 = 0.6–0.8, and the expression of recombinant proteins was induced in the presence of 0.5 mm isopropyl-thio-β-d-galactoside at 37 °C for 4 h. The GST-fused proteins were purified by affinity chromatography on glutathione-Sepharose 4B (GE Healthcare) and eluted with 10 mm reduced glutathione (pH 8.0). Where indicated, protein were released from GST by digestion with the appropriate amount of PreScission protease (GE Healthcare) in digestion buffer (50 mm Tris-HCl, 15 mm NaCl, 1 mm DTT, and 1 mm EDTA, pH 7.5) at 4 °C for 16 h following the manufacturer's instructions.
Preparation of G. duodenalis Proteins
Total soluble proteins were prepared as previously described (3). Briefly, 2 × 109 trophozoites or encysting parasites were collected by chilling on ice and washed 3 times with cold PBS, and the cell pellet was frozen at −70 °C overnight. Cells were resuspended in two volumes of extraction buffer (30 mm Tris-HCl, 1 mm DTT, and 1 mm EDTA, pH 7.4), supplemented with protease-inhibitor mixture (P8340, Sigma) and phosphatase-inhibitor mixture (P2850, Sigma), and then destroyed by sonication. The lysate was centrifuged at 24,000 × g for 30 min at 4 °C, and the supernatant was collected. The protein concentration was measured with the Bradford method (Pierce), and the material was stored at −70 °C.
Western Blot Analysis
Proteins were separated on SDS-PAGE and transferred onto nitrocellulose membrane with 39 mm glycine, 48 mm Tris, 0.1% SDS, and 10% methanol using a semidry apparatus (Bio-Rad). Membranes were blocked with 5% skin milk in T-TBS (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.05% Tween 20) for 1 h and then incubated with the primary antibody (Ab) in T-TBS, 2% skin milk. After incubation with an appropriate HRP-conjugated secondary Ab (1:2000), the interaction was revealed by chemiluminescence (Millipore, Billerica, MA). Mouse M2 anti-FLAG monoclonal Ab (mAb, Sigma) and rabbit anti-α-tubulin mAb (AB-11661, Immunological Sciences) were used at a dilution of 1:500; mouse anti-CWP (Cyst Wall Protein) mAb (1E10) (18) was used 1:20; rabbit anti-g14-3-3 (N14) antiserum (3) was used at 1:5000; mouse anti-HA mAb (HA-7, Sigma) was used at 1:1000; rabbit anti-polyGly and anti-polyGlu polyclonal Abs (pAbs, generous gift of Dr. M. Gorovsky, University of Rochester, NY) were used at 1:2500 and 1:1000, respectively; mouse AXO49 mAb (kindly provided by Dr. M. H. Brè, University of Paris-Sud, France) was used at 1:500.
FLAG Affinity Purification
FLAG-fusion proteins were purified using mouse anti-FLAG M2 mAb covalently bound to agarose beads (Sigma) as previously reported (4). FLAG fusion proteins were directly eluted from the resin by incubation with 200 μm synthetic FLAG-peptide at 4 °C for 1 h. The collected materials were stored at −70 °C until use. For in vitro enzymatic assay of the FLAG-tagged metallopeptidases, proteins bound to the beads were washed 2 times with 50 mm Tris-HCl, pH 7.0, and then eluted with 200 μm synthetic FLAG-peptide in 50 mm Tris-HCl, pH 7.0.
Affinity Purification of g14-3-3 and Mass Spectrometry Analysis
Native g14-3-3 were affinity-purified from G. duodenalis trophozoite total soluble proteins from WBC6 strain or transfected lines using GST-difopein-conjugated beads as previously described (3). Aliquots of g14-3-3 eluted protein were separated on a one-dimensional gel NuPAGE 4–12% (Novex, Invitrogen) run in MOPS buffer and stained with the Colloidal Blue staining kit (Invitrogen). Slices were excised, reduced, alkylated, and digested with modified sequencing-grade trypsin (Promega) as previously described (19). One μl of the digestion supernatant was used for matrix-assisted laser desorption ionization time of flight mass (MALDI-TOF) analysis using the dried droplet technique and 2,5-dihydroxybenzoic acid (Sigma) as matrix. All analyses were performed using a Voyager-DE STR (Applied Biosystems, Framingham, MA) TOF mass spectrometer operated in the delayed extraction mode. All spectra were internally calibrated and processed via the Data Explorer software.
In Vitro Glycylation Assay
20 μl of affinity-purified FLAG-gTTLL3 or control immunopurification from WBC6 strain were mixed with 3 μl of 10× reaction buffer (50 mm MES, pH 7.0, 8 mm MgCl2, 2.5 mm DTT, 5 mm ATP, 25 μm glycine), protease, and phosphatase inhibitors and, where indicated, with 0.25 μg of recombinant Prescission-cleaved g14-3-3 in a final volume of 30 μl. After incubation at 30 °C overnight, the reactions were stopped by adding 10 μl of Laemmli sample buffer and run in 12% SDS-PAGE, transferred on nitrocellulose, and subjected to immunoblot with mouse AXO49 mAb and then with rabbit N14 pAb. The interactions were revealed by chemiluminescence reaction (Millipore).
In Vitro Deglycylation Assay
For the assay, native polyglycylated g14-3-3 was affinity-purified from FLAG-gTTLL3 transgenic line, dialyzed on 50 mm Tris-HCl, pH 7.0, and incubated at 65 °C for 15 min to inactivate any eventual co-purified polyglycylation activity. Eleven μl of affinity-purified FLAG-gDIP1, FLAG-gDIP2, or control immunopurification from WBC6 strain were mixed with 2 μl of 10× buffer (50 mm Tris-HCl, pH 7.5, 1 mm ZnCl2), 200 μg of native polyglycylated g14-3-3, and where indicated with 5 mm EDTA in a final volume of 20 μl. After incubation at 37 °C overnight, the reactions were stopped by adding 10 μl of Laemmli sample buffer and run in 12% SDS-PAGE, transferred on nitrocellulose, and then subjected to immunoblot with rabbit anti-polyGly pAb and mouse anti-HA mAb. After treatment with stripping buffer (100 mm 2-mercaptoethanol, 2% SDS, and 62.5 mm Tris-HCl, pH 6.7) at 50 °C for 30 min, the membrane was probed with rabbit N14 pAb. The interactions were revealed by chemiluminescence (Millipore). The image were acquired and analyzed with the ImageJ 1.43 software.
Parasite Counting
For cell counting, parasites cultured for 3, 6, 12, or 24 h in encysting medium were stained with FITC-conjugated anti-FLAG mAb, Cy3-conjugated anti-CWP mAb, and 4′,6-diamidino-2-phenylindole (DAPI), and microscopy was performed using an Axioplan microscope (Zeiss). For each transgenic parasite line, the total amount of cells spotted on the glass slide and positive for the anti-FLAG staining were counted (∼1000 cells/glass slide). The percentage of trophozoites, encysting parasites, and cysts (both distinguished from trophozoites by co-staining with the anti-CWP mAb) was then calculated. Three independent experiments were performed, and the results were graphed using PRISM 5.0 software (GraphPad Software Inc., San Diego, CA) with S.D.
Confocal Laser Scanning Microscopy
Trophozoites or encysting cells were prepared as previously described (3). Polyclonal rabbit N14 antiserum, rabbit anti-polyGly pAb, monoclonal FITC-conjugated mouse anti-FLAG Ab (Sigma), and the Cy3-conjugated anti-CWP mAb (Waterborne Inc., New Orleans, LA) were used at dilutions of 1:20, and Alexa-Fluor 594-conjugated anti-rabbit secondary Ab (Invitrogen) was used at a 1:500 dilution. After staining, coverslips were extensively rinsed and then mounted on the microscope slide by using Vectashield® mounting medium (Vector Laboratories Inc., Burlingame, CA) containing 300 nm DAPI before confocal laser scanning microscopy analyses. The observations were performed on a Leica TCS SP2 AOBS apparatus, utilizing excitation spectral laser lines at 405, 488, 546, and 594 nm, properly tuned by an acousto-optical tunable filter. The emission wavelengths were selected by a proper setting of the spectral detection system. Image acquisition and processing were conducted by using the Leica Confocal Software (Leica Lasertechnik GmbH, Heidelberg, Germany). Signals from different fluorescent probes were taken in sequential scan mode, and co-localization was detected in yellow. Image processing was performed using the Huygens software (Scientific Volume Imaging BV, Hilversum, The Netherlands). Different fields of view (>200 cells) were analyzed on the microscope for each labeling condition, and representative results are shown.
RESULTS
The Giardial gTTLL3 Is Responsible for the g14-3-3 Polyglycylation in Vivo
The presence of seven giardial ORFs coding for hypothetical proteins containing the TTL domain (20) was confirmed by mining the latest annotated version of G. duodenalis WBC6 genome with the Prosite motif PDOC51221, corresponding to the TTL consensus domain (supplemental Fig. S1A). A phylogenetic analysis conducted on 58 full-length TTLL proteins from unicellular and multicellular eukaryotes showed that gTTLL3 (GL50803_8456) is the only giardial TTLL protein clustering with functional glycylases from other organisms (supplemental Fig. S1B).
To identify the putative enzyme responsible for the glycylation of g14-3-3, the seven gTTLL encoding genes were ectopically expressed in the G. duodenalis parasite as N-terminal FLAG fusion protein under the constitutive promoter of the G. duodenalis α-tubulin gene. Despite several attempts, we were unable to reveal the expression the FLAG-gTTLL7. All the other gTTLL proteins were expressed, and bands of the expected molecular size were detected with an anti-FLAG antibody in the protein lysates from transfected G. duodenalis trophozoites but none in the control WBC6 (Fig. 1A, panel α-FLAG). However, bands at lower molecular size were also visible, especially for FLAG-gTTLL1 and FLAG-gTTLL3, suggesting partial proteolysis of the FLAG-tagged proteins. When protein extracts were tested with the anti-polyGly antibody (20), a band doublet compatible with tubulins (around 50–52 kDa) and a band compatible with g14-3-3 (around 33 kDa) were prominently detected in all the samples, including the control (Fig. 1A, panel α-polyGLY). It is notably that in the lane corresponding to FLAG-gTTLL3 all the positive bands displayed a broader appearance shifted upward, hence suggesting that the ectopic expression of the enzyme could promote an increase in glycylation levels of both proteins. This observation was supported by the immunolabeling with the anti-g14-3-3 serum (Fig. 1A, panel α-14-3-3) that recognized in the FLAG-gTTLL3 sample a broadened band, whereas sharper bands were detected in the other samples. Although the polyglutamylation in G. duodenalis has been reported as a minor modification of α- and β-tubulin (16), we finally tested the reactivity with an anti-polyGlu antibody. A main band at a molecular size compatible with tubulins plus other less intense bands of various molecular size were immunodecorated by the antibody, but no difference in the antibody reactivity was evident among the samples (Fig. 1A, panel α-polyGLU).
FIGURE 1.
In vivo expression and analysis of FLAG-gTTLL proteins in G. duodenalis. A, expression profile of the FLAG-tagged recombinant proteins and their effect on the g14-3-3 is shown. 20 μg of trophozoite protein lysate were separated on 4–12% SDS-PAGE and blotted on a nitrocellulose membrane. The membrane was incubated with the indicated antibodies (bottom). Expected molecular size of the indicated TTLLs is reported on the right of the upper panel. Molecular size markers are on the left. Different time exposition of the same filter incubated with α-FLAG Ab are presented and separated by lines. The equal loading of the sample material is demonstrated by immunostaining with the anti-α-tubulin (panel α-αTUB). B, affinity purification of g14-3-3 is shown. Soluble proteins from trophozoites were incubated with GST-difopein and eluted with 2 mm A8Ap phosphopeptide. An aliquot (1/20) was analyzed in duplicate on 4–12% gradient NuPAGE. Upper panel, a Coomassie-stained gel is shown. Lower panel, immunoblotting with rabbit anti-g14-3-3 serum is shown. Molecular size markers are on the left. The position of the upper and the lower bands, used for MALDI analyses, is indicated. C, mass spectrometry analysis of g14-3-3 is shown. A zoom view of MALDI spectra of g14-3-3 from WBC6 wild type strain and FLAG-gTTL3 transfectant after tryptic digestion of the lower bands (spectra on the left) and of the upper bands (spectra on the right) is shown. m/z ranges have been selected to compare the distribution and the length of polyGly chains. The number of the glycines added to the 230–248 peptide and deduced from the m/z value is reported. The position of the peptide 230–248 is indicated in bold in the g14-3-3 protein sequence (bottom). D, immunopurification of the FLAG-tagged proteins is shown. An aliquot (1/20) of the FLAG peptide-eluted material (WBC6 or FLAG-gTTLL3 transfectant) was separated by 4–12% SDS-PAGE, transferred to a nitrocellulose membrane, and immunostained with α-FLAG mAb. The position of the FLAG-tagged protein is indicated on the right. E, an in vitro glycylation assay is shown. 20 μl of affinity-purified FLAG-gTTLL3 or control immunopurification from WBC6 strain were incubated where indicated with 0.25 μg of recombinant Prescission-cleaved g14-3-3 (Rec. g14-3-3) and separated on 12% SDS-PAGE, transferred on nitrocellulose, and subjected to Western blot with mouse AXO49 Ab (left panel) and then with rabbit α-g14-3-3 serum (right panel). The arrow indicates the molecular size of recombinant g14-3-3. The asterisk on the left indicates the small subunit of the mouse anti-FLAG mAb cross-reacting with the secondary anti-mouse Ab used to detect AXO49.
To confirm that the observed variations in the g14-3-3 electrophoretic profile and anti-polyGly reactivity were ascribable to differences in the polyglycylation level, the native g14-3-3 proteins were purified from control and transfected trophozoites by affinity chromatography with GST-difopein, a synthetic polypeptide that binds 14-3-3 in a phosphorylation-independent manner (Fig. 1B), and analyzed by MALDI-MS. Due to the broad appearance of g14-3-3 in gel, to better appreciate any modification of the polyglycine chain length, each protein band was divided into an upper and lower part, enriched in g14-3-3 with longer and shorter polyglycine chains, respectively, and individual spectra were acquired. The mass spectra of the g14-3-3 protein from the parental WBC6 strain revealed the presence of a peak series at 57-Da intervals compatible with a polyglycylation pattern of the 230DNLNLWVTDSAGDDNAEEK248 (m/z = 2105.92) peptide. This was consistent with the addition of a glycine stretch of 13 up to 30 residues to the 230–248 peptide (Fig. 1C, panels WBC6). The same pattern of peaks was clearly visible also in the mass spectra of the g14-3-3 protein from the FLAG-gTTLL3 transfectant (Fig. 1C, panels FLAG-gTTLL3), although m/z values indicated an increased length of the polyglycine chain, ranging from 14 to 41 residues (Fig. 1C, panels FLAG-gTTLL3). A pattern of peaks comparable with that obtained for the g14-3-3 from the WBC6 strain was observed for all the other transfectants (data not shown).
To finally prove that gTTLL3 was indeed the polyglycylase of g14-3-3, in vitro glycylation activity assays were carried out. Because bacterially expressed GST-gTTLL3 was almost insoluble (data not shown), we use the immunopurified FLAG-gTTLL3 or the WBC6 control immunopurification (Fig. 1D) and, as substrate, the bacterially expressed g14-3-3, which lacks all the post-translational modifications. To evaluate the addition of the polyglycines to the recombinant g14-3-3, we take advantage of the AXO49 antibody developed against the Paramecium axonemal tubulin and able to detect polyglycine chains of at least four residues and also proven to recognize the polyglycylated g14-3-3 (3). As shown in Fig. 1E, the recombinant g14-3-3, properly detected by the anti-g14-3-3 antibody, was recognized by AXO49 only when the protein was incubated in presence of the immunopurified FLAG-gTTLL3. No 14-3-3 signal was observed when the incubation was performed without immunoprecipitated material or with the control WBC6 (Fig. 1E). These results were in an excellent accordance with the phylogenetic position of gTTLL3 together with glycylases from other organisms (Ref. 12 and supplemental Fig. S1).
Two Metallopeptidases of the M20 Family, gDIP1 and gDIP2, Display Deglycylation Activity
In an ongoing proteomic analysis we have identified by mass spectrometry gTTLL3 co-immunoprecipitating with a FLAG-tagged g14-3-3,3 thus supporting the presented functional characterization. Intriguingly, in the same proteomic analysis, tryptic peptides corresponding to two polypeptides annotated as aminoacylhistidine dipeptidases (GL50803_15832 and GL50803_8407), here termed gDIP1 (giardial dipeptidase) and gDIP2, respectively, were also identified. As inferred from the BLAST analysis (data not shown), they shared a high degree of amino acid identity (33–37%) within themselves and with bacterial M20C aminoacylhistidine dipeptidases. According to the peptidase classification on the MEROPS data base (22, 23), these hydrolases belong to the MH clan of metallopeptidases. So far genes coding for M20C peptidases have been identified in the genome of bacteria, archea, and in the protozoans G. duodenalis, Trichomonas vaginalis, and Entamoeba dispar (23). The sequence alignment of gDIP1 and gDIP2 with the Haemophilus somnus aminoacylhistidine dipeptidases and with the prototype carboxypeptidase G2 of Pseudomonas aeruginosa (data not shown) revealed that the essential histidine and carboxyl residues in the metal binding sites were completely conserved (His-94, Asp-137, Glu-168, Asp-191, and His-495 in gDIP1 and His-91, Asp-134, Glu-169, Asp-192, and His-476 in gDIP2) (24), as was the dimerization domain in the central portion of both proteins (25).
Because the carboxypeptidase S (subfamily M20A) of Saccharomyces cerevisiae releases C-terminal aliphatic, aromatic, acid, or basic residues from N-blocked peptides as long as glycine is the penultimate residue (26–29), the possible involvement of gDIP1 and gDIP2 in deglycylation of g14-3-3 was explored. The corresponding genes were cloned and ectopically expressed under the control of an α-tubulin constitutive promoter in G. duodenalis trophozoites as N-terminal FLAG-HA-tagged protein (Fig. 2A, panel α-HA). As shown by an immunoblot with anti-14-3-3 and anti-polyGly, the expression of both FLAG-gDIP1 and FLAG-gDIP2 resulted in the decrease of the molecular size of g14-3-3, compatible either with a proteolytic cleavage of the C terminus of g14-3-3, as the anti-14-3-3 anti-serum recognized the protein N terminus, or more likely, with a shortening of the polyglycine chain, as the anti-polyGly Ab still recognized the protein (Fig. 2A).
FIGURE 2.
In vivo expression and analysis of FLAG-gDIP proteins in G. duodenalis. A, expression profile of the FLAG-HA-tagged gDIPs and their effect on the g14-3-3 protein is shown. Twenty μg of trophozoite protein lysate were separated on 4–12% SDS-PAGE, blotted on nitrocellulose membrane, and probed with the indicated antibodies (on the right). Molecular size markers are on the left. B and C, mass spectrometry analysis of g14-3-3 from WBC6 wild type and FLAG-gTTLL3-, FLAG-gDIP1-, and FLAG-gDIP2-transfected lines are shown. Coomassie bands (B) corresponding to the affinity-purified g14-3-3 were divided in three slices (lower, middle, upper) and independently digested and analyzed by MALDI-ToF (C). Two zoom views of MALDI spectra are shown for the lower band (first two columns on the left) to better visualize the peaks corresponding to the g14-3-3 C-terminal 230–248 peptide with a polyGly chain ranging from 7 to 17 residues. Different zoom views of MALDI spectra have been chosen to optimize the detection of longer glycine chain present in the middle (third column) and upper (fourth column) band for each sample. No polyglycylation peaks pattern were visible in the spectra from the upper bands except for the FLAG-gTTLL3 sample. The number of the glycines added to the 230–248 peptide and deduced from the m/z value are reported.
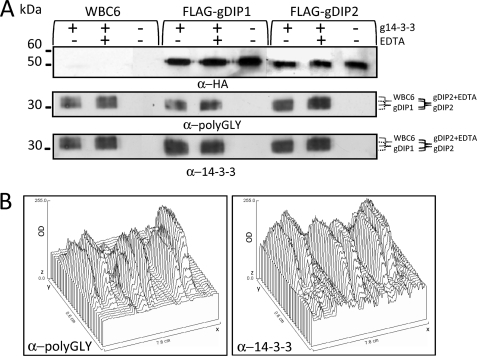
To determine which hypothesis was correct, the endogenous g14-3-3s were purified by GST-difopein affinity from G. duodenalis trophozoites expressing the FLAG-gDIP1 or the FLAG-gDIP2 (Fig. 2B, Coomassie panel) and then analyzed by mass spectrometry. The sequence coverage of the g14-3-3 for all samples was the same (data not shown); the MALDI spectra showed a general reduction in the number of glycines added to the 230DNLNLWVTDSAGDDNAEEK248 peptide(m/z = 2104.92) in the g14-3-3 purified from FLAG-gDIP1 and FLAG-gDIP2 transgenic lines (Fig. 2B, panels FLAG-gDIP1 and FLAG-gDIP2) compared with the wild type WBC6 or with the FLAG-gTTLL3 transgenic line (Fig. 2B). In particular, as can be inferred by molecular mass calculations, the glycines in the WBC6- and in the FLAG-gTTLL3-purified g14-3-3 ranged from 9 to 30 and 10 to 40, respectively (Fig. 2B, panels WBC6 and FLAG-gTTLL3). In contrast, in the FLAG-gDIP1- and FLAG-gDIP2-purified g14-3-3 the number of glycines ranged from 7–8 to 24 (Fig. 2B, panels FLAG-gDIP1 and FLAG-gDIP2). As previously reported, in the WBC6 trophozoite-purified g14-3-3, glycines ranged from 10 to 31 and from 6 to 22 residues in the g14-3-3 purified from 12-h encysting-induced parasite (3). This difference was similar to that observed between wild type WBC6 and FLAG-gDIP1 and FLAG-gDIP2. It is noteworthy that no peak corresponding to the unmodified 230–248 peptide was detected in any of the spectra (data not shown), thus supporting the evidence that complete deglycylation did not occur even in the presence of FLAG-gDIP1 and FLAG-gDIP2 and that the activity of these enzymes as g14-3-3 deglycylases was somehow physiologically limited despite their amount. Phosphorylation of Thr-214 was unaffected even in presence of differential level of polyglycylation, as indicated by the detection of the peak corresponding to the phosphorylated peptide 202–219 in the MALDI spectra in all the samples (data not shown), whereas the unmodified 202–219 peptide was never observed. Taking advantage of the possibility to easily obtain endogenous g14-3-3 by affinity purification with GST-difopein, an in vitro deglycylation assay was carried out using the g14-3-3 from the FLAG-gTTLL3-transfected line as the reaction substrate. At first we tried to perform the assay with bacterially expressed GST-gDIP1 and GST-gDIP2; however, the recombinant proteins were found to be enzymatically inactive in all the conditions tested (data not shown). We then succeeded in performing this assay using the immunopurified FLAG-tagged gDIP1 or gDIP2 expressed in G. duodenalis trophozoites. When the polyglycylated g14-3-3 was incubated in the presence of immunopurified FLAG-tagged gDIP1 or gDIP2, the apparent molecular size of g14-3-3 showed a slight decrease mainly in the FLAG-gDIP1 sample, as compared with the g14-3-3 in the WBC6 control (Fig. 3A). Because metallopeptidases are generally inhibited by metal chelating agents, the assay was performed also in the presence of EDTA, and under our experimental conditions, the activity of FLAG-gDIP2, but not of FLAG-gDIP1, was partially inhibited (Fig. 3A). These observed shifts of the molecular size of g14-3-3 were more evident by the densitometric analysis of the immunoblots (Fig. 3B).
FIGURE 3.
In vitro deglycylation assay. A, 20 μl of FLAG affinity-purified gDIP1 and gDIP2 or control immunopurification from the WBC6 strain were incubated as indicated on the top of the panel with 0.2 μg of difopein affinity-purified g14-3-3 from FLAG-gTTLL3-transfected trophozoite in the presence or not of EDTA and separated on 12% SDS-PAGE, transferred on nitrocellulose, and subjected to Western blot with mouse α-polyGly Ab (middle panel) and then with rabbit-g14-3-3 serum (lower panel) and α-HA Ab (upper panel). The brackets (right side) indicate the molecular size range of the polyglycylated g14-3-3; incubation with WBC6 IP (gray bracket) and with FLAG-DIP1 immunoprecipitate (dotted bracket) with or without EDTA and incubation with FLAG-DIP2 immunoprecipitate (black bracket) and with FLAG-DIP2 with EDTA (empty bracket). Molecular size markers are on the left. B, densitometric analysis of the Western blots with α-polyGly and α-g14-3-3 presented in panel A was performed with ImageJ software. In the graphs the peaks represent the optic density (OD) of the immunostained bands reported as arbitrary value (z axes) and the position of the peaks (in cm) correspond to the position of the bands on the membrane (x and y axis). The molecular size shift of polyglycylated g14-3-3 as a consequence of the incubation with FLAG-gDIPs in the presence or absence of EDTA is more evident.
Expression of FLAG-gDIP1 and of FLAG-gDIP2, but Not of FLAG-gTTLL3, Affects the Encystation Process
Because the different polyglycylation status of g14-3-3 has been implicated in the intracellular localization of the protein (3, 4) as well as in the regulation of parasite encystation timing (4), the effect of FLAG-tagged gTTLL3, gDIP1, and gDIP2 expression during G. duodenalis encystation was further investigated. Because a puromycin can increase levels of CWPs and cyst formation in vegetative cell cultures (30), as a further control we used a transgenic WBC6 transfect with the empty pTUB-FLAGpac vector (WBC6 + vector). Expression of transgenes did not produce any observable effect on trophozoite growth compared with that of the WBC6 or the WBC6 + vector (data not shown). Immunostaining with an anti-FLAG mAb clearly demonstrated that the FLAG-tagged proteins are well expressed at 12 h after the encystation induction (Fig. 4A, panel α-FLAG, 12 h). As shown by immunostaining with anti-g14-3-3 and anti-polyGly (Fig. 4A), the molecular size reduction of g14-3-3 during encystation, consistent with the polyglycine chain shortening (Lalle et al. (3)), occurred in the FLAG-gTTLL3-expressing parasite as well as in the control WBC6 and the WBC6 + vector. These results indicated that either the enzymatic activity of FLAG-gTTLL3 was inhibited or deglycylases were activated during the encystation. On the contrary, in both the FLAG-gDIP1 and FLAG-gDIP2 transgenic lines the molecular size of g14-3-3 at the trophozoite stage was close to that of the protein in the WBC6 and the WBC6 + vector at 12 h after the encystation induction (Fig. 4A, α-g14-3-3 panel). Notably, no further molecular size reduction of g14-3-3 was observed in the FLAG-gDIP1 and FLAG-gDIP2 samples at 12 h. Furthermore, g14-3-3 was still recognized by the anti-polyGly Ab, indicating that deglycylation of the protein was not complete. These observations provided evidence that the expression of each FLAG-tagged dipeptidase is sufficient by itself to promote in vivo deglycylation of g14-3-3 to a level resembling the physiological process observed in encysting G. duodenalis parasites (3).
FIGURE 4.
Effect of FLAG-tagged gTTLL3 and gDIPs during G. duodenalis encystation. A, for Western blot analysis, 30 μg of total soluble proteins extracted from transfected trophozoites (T) or encysting parasite (12 h) from transfected lines (FLAG-gDIP1, FLAG-gDIP2, and FLAG-gTTLL3) from the control WBC6 strain or from the WBC6 line transfected with empty pTUB-FLAGpac plasmid (WBC6 + vector) were used in each lane. Proteins were separated on a 12% SDS-PAGE and transferred onto a PVDF membrane and probed with the indicated antibodies. The arrows on the right of the α-polyGLY panel indicate the tubulin and g14-3-3 bands. B, in vivo effect of FLAG-tagged gTTLL3 and gDIPs on encystation process is shown. The amount of trophozoites, encysting parasites, and cysts expressing the FLAG-tagged proteins were estimated by co-staining with anti-CWP and anti-FLAG antibodies and counted in three independent experiments after 3, 6, 12, and 24 h of growth in encysting medium. Results were bar-graphed as the percentage of the total amount of cells observed (∼1000 parasites per experiment/transfected line). The error, expressed as S.D., is reported. C, quantitative real-time PCR analysis of gttll3, gdip1, gdip2, and cwp1 genes amplified with gene-specific primers at different time points during the encystation (0, 3, 6, and 12 h) is shown. The threshold cycle of all genes was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (gap1). Relative gene expression was calculated using the 2−ΔΔCt method.
Moreover, despite comparable amounts of α-tubulin detected in all samples (Fig. 4A, α-αTUB panel), the anti-polyGly Ab, besides the g14-3-3, strongly recognized a band compatible with tubulin in the FLAG-gTTLL3 trophozoites and, with lower intensity, in the WBC6 and the WBC6 + vector trophozoites, but not in the 12-h samples (Fig. 4A, α-polyGLY panel), thus suggesting a possible effect of FLAG-gTTLL3 also in the tubulin glycylation. No staining at the molecular size of tubulin was observed with the anti-polyGly Ab in any of the FLAG-gDIP1 and FLAG-gDIP2 samples (Fig. 4A, panel α-polyGLY).
Surprisingly, when the same samples were assayed for CWP expression levels, the proteins were almost undetectable in the 12-h-induced sample from the FLAG-gDIP2 transgenic line (Fig. 4A, panel α-CWP) but readily detectable in the corresponding samples from WBC6, WBC6 + vector, FLAG-gTTLL3, and FLAG-gDIP1. As expected, the protein(s) was not detectable in all trophozoite samples.
To correlate the impaired expression of CWP proteins observed in the FLAG-gDIP2 line with the cyst development, the number of trophozoites, encysting parasites, and cysts present in the G. duodenalis cultures were counted at different time points during growth in encysting medium. Encysting parasites and cysts were distinguished (Fig. 4B) based on the expression of the CWP protein. In comparison with the WBC6 + vector control line, the in vitro differentiation of trophozoites into cysts was strongly hampered in the FLAG-gDIP2 and, to a lesser extent, in FLAG-gDIP1 line (Fig. 4B). In particular, in the FLAG-gDIP2 line the effect was already evident at 3 h. At this time point, no encysting parasites were detectable compared with 5% in the WBC6 + vector control line. At 12 h no cysts were visible yet, and only 15% of encysting parasites were counted in the FLAG-gDIP2 line compared with 9% of cysts and 35% of encysting parasites in the control. After 24 h, the control contained more than 45% cysts, in contrast with less than 5% in the FLAG-gDIP2 line. However, the latter cysts were not stained by the anti-FLAG mAb, indicating that the expression of FLAG-gDIP2 was somehow abolished. A similar but milder effect was observed with FLAG-gDIP1 transfectants. Cysts (around 1%) appeared at 12 h and reached 20% at 24 h, whereas starting from 6 h after the induction of encystation, the percentage of encysting parasites was comparable with the control. On the contrary, parasites expressing the FLAG-gTTLL3 showed an encystation profile comparable with that of the control (Fig. 4B), further supporting the previous observation that expression of FLAG-gTTLL3 did not impair the physiological deglycylation of g14-3-3 nor, possibly, that of tubulins.
To give an explanation for the adverse effect of FLAG-gDIP2 expression on the development of G. duodenalis cysts, the gene expression profile of gdip1, gdip2, and gttll3 was investigated in the parental WBC6 strain at different time points after the induction of encystation (Fig. 4C). The relative gene expression level of gdip2, normalized against the constitutive gap1 (glyceraldehyde-3-phosphate dehydrogenase) gene, decreased 2-fold from 3 to 12 h after encystation induction, whereas the expression level of gdip1 was almost constant throughout. The expression level of gttll3 gene was up-regulated 2.5-fold at 3 h, compared with the trophozoite stage (0 h) but subsequently decreased at 6 and 12 h post-induction. As expected, the expression of the encystation-induced cwp1 gene was strongly up-regulated, starting from 3 h post-induction.
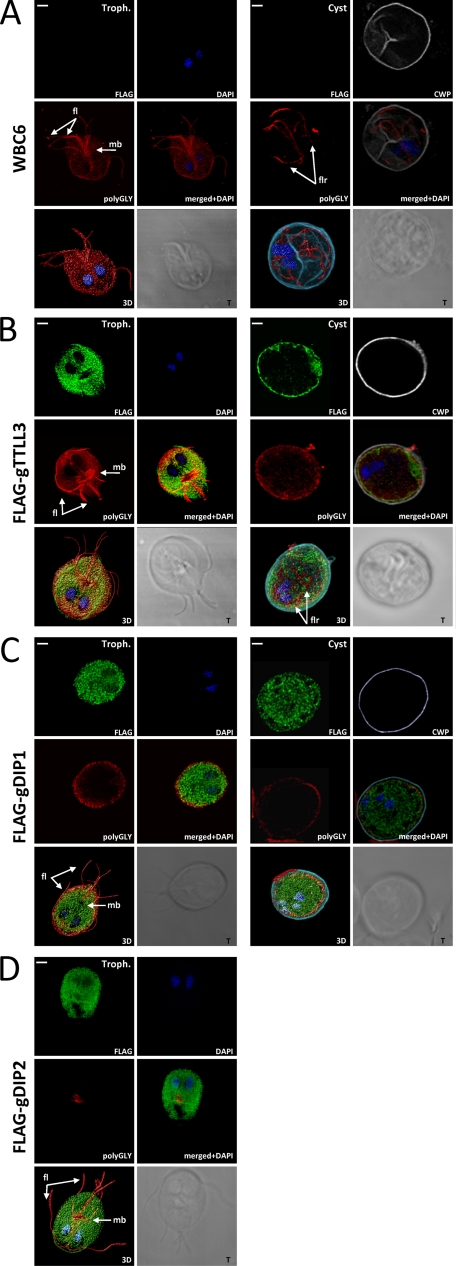
Intracellular Localization of FLAG-gTTLL3 and FLAG-gDIPs and Their Effect on g14-3-3 Localization and on the Overall Polyglycylation Levels
The shortening of the g14-3-3 polyglycine tail during G. duodenalis encystation coincides with the nuclear translocation of the protein (3), and the absence of a polyglycylation tail promotes the constitutive localization of a g14-3-3 mutant (E246A) to the nuclei (4). The intracellular localization of FLAG-gTTLL3, FLAG-gDIP1, and FLAG-gDIP2 and the effects of their overexpression on g14-3-3 compartmentalization were studied by confocal laser scanning microscopy in the G. duodenalis trophozoite, encysting, and cyst stages. In parasites expressing the FLAG-gTTLL3 (Fig. 5A), the anti-FLAG mAb stained the cell body at all stages and also the periphery of the cyst. No staining of other subcellular structures as well as nuclei was detected. A partial overlapped staining (yellow color) between the anti-14-3-3 and the FLAG signals was evident in the cytoplasm of all stages (Fig. 5A). However, no co-localization at the cell periphery was visible in the cyst (Fig. 5A). The expression of FLAG-gTTLL3 did not affect the nuclear localization of 14-3-3, which was excluded from the nuclei of trophozoites and cysts but localized in the nuclei of encysting parasites (Fig. 5A). These data are in agreement with the observed deglycylation of g14-3-3 during encystation, even when FLAG-gTTLL3 is expressed.
FIGURE 5.
Subcellular localization of the endogenous g14-3-3 in the FLAG-tagged gTTLL3 and gDIP-transfected G. duodenalis parasites. Parasites transfected with FLAG-gTTLL3 (A), FLAG-gDIP1 (B), and FLAG-gDIP2 (C) are shown. Confocal laser scanning microscopy observations of trophozoites (Troph.), encysting parasites (at 12 h post-encystations induction (12-h Encyst.)), and cysts stained with FITC-conjugated anti-FLAG mAb (shown in green) with anti-g14-3-3 serum are revealed by Alexa Fluor-594 goat anti-rabbit Ab (red), with Cy3-conjugated anti-CWP mAb (pseudocolor gray), and with DAPI (blue). Displayed micrographs correspond to the single stack encompassing the center of the two nuclei of the trophozoite or the encysting parasite or at least two of the four cyst nuclei. merged+DAPI, merged images with DAPI-stained nuclei; 3D, three-dimensional reconstructions of the complete stack series for each acquisition; T, transmission light acquisition. Scale bars, 2 μm. Arrows indicate the position of nuclei (N), median body (mb), marginal groove (mg), and ventrolateral flanges (vf) (48).
In parasites expressing the FLAG-gDIP1 (Fig. 5B), the FLAG-tagged protein localized in the cytoplasm of all parasite stages as well as in the nuclei of trophozoites and encysting parasites and in the median body of encysting parasites. Similarly to what reported for the FLAG-gTTLL3 transgenic line, the localization of g14-3-3 was not affected by the FLAG-gDIP1 expression at any parasite stage (Fig. 5B).
Finally, in parasites expressing the FLAG-gDIP2 (Fig. 5C), no cyst with anti-FLAG staining was found, in accordance with the observed difficulty of this transgenic line to form cysts. In the trophozoite stage, anti-FLAG markedly stained the cell ventral surface, nearly corresponding to specialized contractile structures with a role in G. duodenalis attachment and termed marginal groove and ventrolateral flanges (31–33). A diffuse anti-FLAG staining was also present in the nuclei and in the median body (Fig. 5C). Similar staining was evident in the few parasites that were induced for encystation (Fig. 5C). Notably, the forced expression of FLAG-gDIP2 strongly affected g14-3-3 localization; already at the trophozoite stage, the protein was detectable in the nuclei of trophozoites and encysting parasites and also accumulated in the area corresponding to the median body (Fig. 5C).
We then studied the overall effect of the FLAG-tagged protein expression on the staining profile of G. duodenalis cells by the anti-polyGly Ab. In the untransfected WBC6 parasites (Fig. 6A and supplemental Fig. 2), the anti-polyGly Ab markedly stained the cell body, the flagella, and the median body in trophozoites, whereas in encysting parasites the staining intensity slightly decreased (more evident in the three-dimensional reconstruction panel), and in cysts only the flagellar remnants were visible. Our observations are partially in agreement with previous immunofluorescence studies of G. duodenalis trophozoites using the AXO49 Ab, which was used only to study polyglycylated tubulin (17). A similar but more intense staining was evident in parasites expressing the FLAG-gTTLL3 (Fig. 6B and (supplemental Fig. 2). In particular, in the trophozoite, the median body appeared enlarged and massively stained (Fig. 6B) when compared with the WBC6 (Fig. 6A). In all FLAG-gDIP1-expressing parasite stages (Fig. 6C and supplemental Fig. 2), the anti-polyGly Ab immunodecorated the flagella and faintly stained the cell body, whereas it never marked the median bodies. Intriguingly, the periphery of the cyst was stained by the anti-polyGly, but the flagellar remnants were not detectable (Fig. 6C). On the contrary, in trophozoites expressing the FLAG-gDIP2- (Fig. 6D), the anti-polyGly b labeling of the cell body was almost undetectable, whereas the flagella and the median body were stained. A similar pattern was observed in the encysting parasite (supplemental Fig. S2).
FIGURE 6.
Immunofluorescence staining of WBC6- and FLAG-tagged gTTLL3- and gDIPs-transfected G. duodenalis parasites with anti-polyGly antibody. Wild type parasite WBC6 (A) and parasites transfected with FLAG-gTTLL3 (B), FLAG-gDIP1 (C), and FLAG-gDIP2 (D) are shown. Confocal laser scanning microscopy observations of trophozoites (Troph.) and cysts stained with FITC-conjugated anti-FLAG mAb (shown in green), with anti-polyGly Ab revealed by Alexa Fluor-594 goat anti-rabbit Ab (red), with Cy3-conjugated anti-CWP mAb (pseudocolor gray), and with DAPI (blue) are shown. Displayed micrographs correspond to the single stack encompassing the center of the two nuclei of trophozoite or encysting parasite or at least two of the four cyst nuclei. merged+DAPI, merged images with DAPI-stained nuclei; 3D, three-dimensional reconstructions of the complete stack series for each acquisition; T, transmission light acquisition. Scale bars, 2 μm. Arrows indicate the position of median body (mb), flagella (fl), and flagella remnants (flr).
DISCUSSION
In this work we have shown that in G. duodenalis, despite the presence of several TTLL encoding genes, only the gTTLL3 protein displays an appreciable polyglycylation activity on g14-3-3. An in vitro glycylation assay using the AXO49 Ab, which recognizes polyglycine chains of at least four residues (3, 34), together with the recombinant unmodified g14-3-3 suggests that gTTLL3 could be initiating/elongating enzymes that catalyze both the addition of the first glycine on the C terminus of g14-3-3 and the subsequent elongation of the polyglycine chain. Recent findings have demonstrated that in vertebrates polyglycylation of microtubules occurs by a two-step mechanism requiring the combined activities of the TTLL3/8 and TTLL10 enzymes (9, 10). A similar mechanism probably functions in the ciliate T. thermophila, where the TTLL3 homolog shows only glycine-ligase/initiase activity on tubulin (15). On the contrary, in organisms lacking TTLL10 homologs, bifunctional activities have been reported. In D. melanogaster, DmTTLL3A mono- and polyglycylates the α- and the β-tubulin, whereas DmTTLL3B mono- and polyglycylates non-tubulin proteins (10). Notably, mammalian TTLL10 is also able to act as a bifunctional enzyme on nucleosome assembly proteins (8). The evidence that gTTLL3 is the only member of the giardial TTLL family clustering close to other mono- and polyglycylases and that G. duodenalis does not have a TTLL10 homolog protein (Ref. 20; supplemental Fig. S1) is in favor of a direct role of gTTLL3 not only in g14-3-3 but most probably also in the tubulin polyglycylation.
As suggested by anti-polyGly immunoblotting and immunolocalizations, expression of the FLAG-gTTLL3 led also to an increase in polyglycylation levels of tubulin and microtubule structures. However, further studies are needed to clarify the activity of gTTLL3 on G. duodenalis tubulin as well as to define the function of the other gTTLLs, in particular gTTLL7, the only one that failed to be expressed.
In T. thermophila, TTLL3 protein homologs localize primarily in cilia or basal bodies, with the exception of TTLL3Dp, which is present in the cell body (15). Here, we show that the FLAG-gTTLL3 protein partially co-localizes with g14-3-3 at all stages, in agreement with its polyglycylation activity toward g14-3-3, and it is not present in flagella or basal bodies. Intriguingly, in cysts the FLAG-gTTLL3 signal accumulates just underneath the cyst wall, most probably at the plasmalemma level. In agreement with gene expression data, this peculiar localization could suggest that physiologically the protein is constitutively expressed and stored during the dormant cyst stage to be eventually available during excystation. A similar conservative strategy is adopted by G. duodenalis during cyst formation; the ventral disc and flagella are broken into several large fragments, stored in the cytoplasm of the cyst, and apparently reassembled during excystation (35, 36).
The production of FLAG-gTTLL3 protein did not affect the deglycylation of g14-3-3 during encystation, g14-3-3 localization, or the encystation timing. These data suggest that the developmental modulation of polyglycine chain length is exerted by deglycylating enzymes irrespective of the starting polyglycylation level. This hypothesis is supported by the almost constant expression of the gttll3 gene in the wild type G. duodenalis strain.
To the best of our knowledge this is the first study reporting the identification of two metallopeptidases, gDIP1 and gDIP2, with a deglycylase activity. The reversible nature of polyglycylation and the existence of tubulin deglycylases were previously shown in cytoplasmic extracts of the ciliate Paramecium tetraurelia, but the enzymes were not identified (6). gDIP1 and gDIP2 belong to the MH clan, subfamily M20C. Metallopeptidases of the clan MH contains six families (M18, M20, M25, M28, M40, and M42) with enzymes having known activities such as carboxypeptidases or aminopeptidases. Members of the M20 metallohydrolase family, further divided into four subfamilies (A–D), require co-catalytic divalent metal ions, generally zinc, for their activity, and all include different exopeptidases, like the members of the clans MF and MG (37). However, the M20 and M28 families are unique in attacking the C terminus of the substrate (37). The glutamate carboxypeptidase G2 of Pseudomonas sp. (subfamily M20A) hydrolyzes the C-terminal glutamate moiety from folic acid and its analogues (38), the tripeptidase T (M20B) of Lactococcus lacti hydrolyzes only tripeptides (39), the bacterial aminoacylhistidine dipeptidase performs hydrolysis of dipeptides (40), and the thermostable carboxypeptidase Ss1 (M20D) of the archean Sulfolobus solfataricus is able to cleave N-blocked tripeptides (41).
The ectopic expression of FLAG-gDIP1 and FLAG-gDIP2 in G. duodenalis results in a remarkable reduction in the length of the polyglycine chain of g14-3-3, down to a size comparable with that observed in the g14-3-3 from wild type encysting parasites (3). Nevertheless, a complete deglycylation of g14-3-3 was not observed at any stage in FLAG-gDIP1 or FLAG-gDIP2 transgenic parasite. A possible explanation for these observations might be related to the need for FLAG-gDIP1 and -2 activities for a cofactor protein, likely present in limiting amounts. Alternatively or in addition, the function of both enzymes might be partially counteracted by the activity of endogenous gTTLL3. The physiological, simultaneous expression of both the gttll3 and gdip1 genes in trophozoites and encysting parasites weighs in favor of the second hypothesis, again indicating that in G. duodenalis a fine balance exists between polyglycylation and deglycylation of g14-3-3. However, even though the partial deglycylation of g14-3-3 obtained in vitro with FLAG-tagged enzymes confirms the in vivo observations, the experimental conditions used in vitro might not allow the enzymes to express their full activities. Furthermore, the modest-to-absent inhibitory effect exerted by the metal chelator EDTA on the FLAG-gDIPs activities has also been reported for other metallopeptidases of different clans, such as human aminopeptidase N, Bacillus stearothermophilus aminopeptidase I, and Sabellastarte magnifica metallocarboxypeptidase (42–44). In these cases the absence of the inhibitory effect has been related to a strong binding of the metal to the enzyme.
Previously, we have demonstrated that the E246A mutant of g14-3-3, which cannot be glycylated, localizes to the nuclei already at the trophozoite stage, resulting in a faster differentiation of trophozoites into cysts (4). Here, we show that despite the comparable shortening of g14-3-3 polyglycine chain attained by both metallopeptidases, the g14-3-3 localization in the trophozoites nuclei occurs only in FLAG-gDIP2 transfectants. Moreover, we observe that the expression of FLAG-gDIP2 strongly hampers cyst formation and affects CWP protein levels, whereas the forced expression of FLAG-gDIP1 results only in a limited reduction of encystation efficiency. Nevertheless, these results are not in conflict, as the g14-3-3 E246A point mutation only affects the g14-3-3 itself, whereas the expression of FLAG-gDIP1 and FLAG-gDIP2 might exert its effect on multiple targets, most likely including tubulin. Taken together, these data indicate that gDIP1 and gDIP2, besides their role in deglycylation of 14-3-3 and possibly tubulin, might carry out other functions, as also suggested by the different localization of the two proteins. Indeed, FLAG-gDIP2 labeling is evident in the median body, in the marginal groove, and in the ventrolateral flanges, whereas FLAG-gDIP1 localizes also in the nuclei. Moreover, the fact that gdip2 is down-regulated at the encystation, whereas gdip1 is almost constitutively expressed supports the possible involvement of these enzymes in different cellular processes. Deglycylation of nuclear proteins, such as transcriptional or chromatin remodeling factors, might be responsible for the defect in encystation most clearly observed in the FLAG-gDIP2 transgenic line. It has been proposed that glycylation by TTLL10 of nucleosome assembly protein 1 (NAP1) in the mouse might modulate NAP1-histone interactions, thus inducing transcriptional repression during spermiogenesis (8). In D. melanogaster, depletion of DmTTLL3B protein results in several defects attributed to the potential lack of glycylation of its substrates, including chaperonins and motor proteins (10). Intriguingly, G. duodenalis motor proteins are localized in an area corresponding to the lateral crest of the trophozoite (45). However, further studies are needed to verify the existence of other polyglycylation substrates and their possible involvement in the differentiation of G. duodenalis.
Recently, Kimura et al. (46) published the identification in Caenorhabditis elegans and humans of tubulin deglutamylases as cytosolic carboxypeptidases of the MC clan, M14 family, subfamily M14D of metallopeptidases. G. duodenalis has four genes encoding proteins assigned to the M14 family but all classified as non-peptidase homologs (23). Notably, metallopeptidases of the MC and the MH clans share no detectable sequence homologies (47), thus, excluding the hypothesis of a common evolutionary origin. However, they show a common protein fold similar to peptidases in which two zinc atoms are essential for their activity (such as MH metallopeptidases), implying that metallopeptidase activity might be acquired in independent evolutionary events (37). Because the identified deglutamylases (46) and deglycylases (this study) do not seem to belong to a single well conserved protein family, as in the case of the TTLLs, but share metallopeptidase properties, it is plausible that other metallopeptidases of different clans might exert similar functions in other organisms. Indeed, members of the metallopeptidase clans M14 and M28 and subfamilies M20A and M20D, but none of the M20C, have been identified in the partially annotated genomes of the ciliates T. thermophila and P. tetraurelia as well as in humans and D. melanogaster (23). In conclusion, we have identified in G. duodenalis a polyglycylase and two deglycylases that act in concert to modulate the stage-dependent glycylation state of the multifunctional regulatory g14-3-3 protein.
Supplementary Material
Acknowledgments
We thank Dr. Maria C. Touz, (Instituto de Investigación Medica Mercedes y Martín Ferreyra, Consejo Nacional de Investigaciones Científicas y Técnicas, Cordoba, Argentina) for the kind gift of the pTubApaH7-HApac vector, Dr. Gorowky (University of Rochester, NY) for the anti-poly glutamine and anti-polyglycine antibodies, and Dr. Mariè-Helen Bré (University of Paris-Sud, France) for the AXO49 antibody. Mass spectrometry experiments were carried out by the Telethon Proteomic Service (GTF08002).
This work was supported in part by European Commission Contract SANCO/2006/FOODSAFETY/032.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and supplemental Tables 1 and 2 and Figs. S1 and S2.
M. Lalle, S. Camerini, S. Cecchetti, A. Sayadi, M. Crescenzi, and E. Pozio, manuscript in preparation.
- TTLL
- tubulin tyrosine ligase (TTL)-like
- g-
- giardial
- Ab
- antibody
- pAb
- polyclonal Ab
- CWP
- cyst wall protein.
REFERENCES
- 1. Thompson R. C. (2000) Int. J. Parasitol. 30, 1259–1267 [DOI] [PubMed] [Google Scholar]
- 2. Morrison H. G., McArthur A. G., Gillin F. D., Aley S. B., Adam R. D., Olsen G. J., Best A. A., Cande W. Z., Chen F., Cipriano M. J., Davids B. J., Dawson S. C., Elmendorf H. G., Hehl A. B., Holder M. E., Huse S. M., Kim U. U., Lasek-Nesselquist E., Manning G., Nigam A., Nixon J. E., Palm D., Passamaneck N. E., Prabhu A., Reich C. I., Reiner D. S., Samuelson J., Svard S. G., Sogin M. L. (2007) Science 317, 1921–1926 [DOI] [PubMed] [Google Scholar]
- 3. Lalle M., Salzano A. M., Crescenzi M., Pozio E. (2006) J. Biol. Chem. 281, 5137–5148 [DOI] [PubMed] [Google Scholar]
- 4. Lalle M., Bavassano C., Fratini F., Cecchetti S., Boisguerin P., Crescenzi M., Pozio E. (2010) Int. J. Parasitol. 40, 201–213 [DOI] [PubMed] [Google Scholar]
- 5. Aitken A. (2006) Semin. Cancer. Biol. 16, 162–172 [DOI] [PubMed] [Google Scholar]
- 6. Bré M. H., Redeker V., Vinh J., Rossier J., Levilliers N. (1998) Mol. Biol. Cell 9, 2655–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vinh J., Langridge J. I., Bré M. H., Levilliers N., Redeker V., Loyaux D., Rossier J. (1999) Biochemistry 38, 3133–3139 [DOI] [PubMed] [Google Scholar]
- 8. Ikegami K., Horigome D., Mukai M., Livnat I., MacGregor G. R., Setou M. (2008) FEBS Lett. 582, 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikegami K., Setou M. (2009) FEBS Lett. 583, 1957–1963 [DOI] [PubMed] [Google Scholar]
- 10. Rogowski K., Juge F., van Dijk J., Wloga D., Strub J. M., Levilliers N., Thomas D., Bré M. H., Van Dorsselaer A., Gaertig J., Janke C. (2009) Cell 137, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 11. Ersfeld K., Wehland J., Plessmann U., Dodemont H., Gerke V., Weber K. (1993) J. Cell Biol. 120, 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janke C., Rogowski K., Wloga D., Regnard C., Kajava A. V., Strub J. M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., Suryavanshi S., Gaertig J., Eddé B. (2005) Science 308, 1758–1762 [DOI] [PubMed] [Google Scholar]
- 13. van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. (2007) Mol. Cell 26, 437–448 [DOI] [PubMed] [Google Scholar]
- 14. Wloga D., Rogowski K., Sharma N., Van Dijk J., Janke C., Eddé B., Bré M. H., Levilliers N., Redeker V., Duan J., Gorovsky M. A., Jerka-Dziadosz M., Gaertig J. (2008) Eukaryot. Cell 7, 1362–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wloga D., Webster D. M., Rogowski K., Bré M. H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S. T., Gaertig J. (2009) Dev. Cell 16, 867–876 [DOI] [PubMed] [Google Scholar]
- 16. Weber K., Schneider A., Westermann S., Müller N., Plessmann U. (1997) FEBS Lett. 419, 87–91 [DOI] [PubMed] [Google Scholar]
- 17. Campanati L., Bré M. H., Levilliers N., de Souza W. (1999) Biol. Cell 91, 499–506 [DOI] [PubMed] [Google Scholar]
- 18. Winiecka-Krusnell J., Linder E. (1995) Eur. J. Clin. Microbiol. Infect. Dis. 14, 218–222 [DOI] [PubMed] [Google Scholar]
- 19. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 20. Redeker V., Levilliers N., Vinolo E., Rossier J., Jaillard D., Burnette D., Gaertig J., Bré M. H. (2005) J. Biol. Chem. 280, 596–606 [DOI] [PubMed] [Google Scholar]
- 21. Deleted in proof.
- 22. Barrett A. J., Tolle D. P., Rawlings N. D. (2003) Biol. Chem. 384, 873–882 [DOI] [PubMed] [Google Scholar]
- 23. Rawlings N. D., Barrett A. J. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., pp. 231–268, Elsevier, London [Google Scholar]
- 24. Neuwald A. F., Liu J. S., Lipman D. J., Lawrence C. E. (1997) Nucleic Acids Res. 25, 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rowsell S., Pauptit R. A., Tucker A. D., Melton R. G., Blow D. M., Brick P. (1997) Structure 5, 337–347 [DOI] [PubMed] [Google Scholar]
- 26. Spormann D. O., Heim J., Wolf D. H. (1991) Eur. J. Biochem. 197, 399–405 [DOI] [PubMed] [Google Scholar]
- 27. Bordallo J., Bordallo C., Gascón S., Suárez-Rendueles P. (1991) FEBS Lett. 283, 27–32 [DOI] [PubMed] [Google Scholar]
- 28. Félix F., Brouillet N. (1966) Biochim. Biophys. Acta 122, 127–144 [PubMed] [Google Scholar]
- 29. Suarez-Rendueles P., Bordallo J. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., pp. 950–951, Elsevier, London [Google Scholar]
- 30. Su L. H., Lee G. A., Huang Y. C., Chen Y. H., Sun C. H. (2007) Mol. Biochem. Parasitol. 156, 124–135 [DOI] [PubMed] [Google Scholar]
- 31. Friend D. S. (1966) J. Cell Biol. 29, 317–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feely D. E., Schollmeyer J. V., Erlandsen S. L. (1982) Exp. Parasitol. 53, 145–154 [DOI] [PubMed] [Google Scholar]
- 33. Sousa M. C., Gonçalves C. A., Bairos V. A., Poiares-Da-Silva J. (2001) Clin. Diagn. Lab. Immunol. 8, 258–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bré M. H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J. M., Rossler J., Johnson T., Adoutte A., Levilliers N. (1996) J. Cell Sci. 109, 727–738 [DOI] [PubMed] [Google Scholar]
- 35. Palm D., Weiland M., McArthur A. G., Winiecka-Krusnell J., Cipriano M. J., Birkeland S. R., Pacocha S. E., Davids B., Gillin F., Linder E., Svärd S. (2005) Mol. Biochem. Parasitol. 141, 199–207 [DOI] [PubMed] [Google Scholar]
- 36. Midlej V., Benchimol M. (2009) Parasitol. Int. 58, 72–80 [DOI] [PubMed] [Google Scholar]
- 37. Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Minton N. P., Atkinson T., Sherwood R. F. (1983) J. Bacteriol. 156, 1222–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mori S., Nirasawa S., Komba S., Kasumi T. (2005) Biochim. Biophys. Acta 1748, 26–34 [DOI] [PubMed] [Google Scholar]
- 40. Henrich B., Klein J. R. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., pp. 951–953, Elsevier, London [Google Scholar]
- 41. Colombo S., Toietta G., Zecca L., Vanoni M., Tortora P. (1995) J. Bacteriol. 177, 5561–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moser P., Roncari G., Zuber H. (1970) Int. J. Protein. Res. 2, 191–207 [PubMed] [Google Scholar]
- 43. Vanderheyden P. M., Demaegdt H., Swales J., Lenaerts P. J., De Backer J. P., Vogel L. K., Vauquelin G. (2006) Fundam. Clin. Pharmacol. 20, 613–619 [DOI] [PubMed] [Google Scholar]
- 44. Alonso-del-Rivero M., Trejo S. A., Rodríguez de la Vega M., González Y., Bronsoms S., Canals F., Delfín J., Diaz J., Aviles F. X., Chávez M. A. (2009) FEBS J. 276, 4875–4890 [DOI] [PubMed] [Google Scholar]
- 45. Narcisi E. M., Paulin J. J., Fechheimer M. (1994) J. Parasitol. 80, 468–473 [PubMed] [Google Scholar]
- 46. Kimura Y., Kurabe N., Ikegami K., Tsutsumi K., Konishi Y., Kaplan O. I., Kunitomo H., Iino Y., Blacque O. E., Setou M. (2010) J. Biol. Chem. 285, 22936–22941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wouters M. A., Husain A. (2001) J. Mol. Biol. 314, 1191–1207 [DOI] [PubMed] [Google Scholar]
- 48. Elmendorf H. G., Dawson S. C., McCaffery J. M. (2003) Int. J. Parasitol. 33, 3–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.