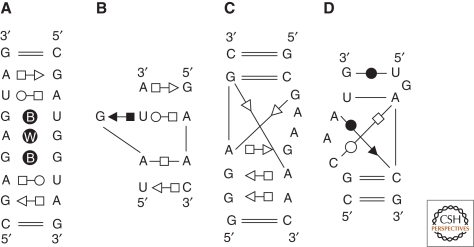
Figure 1.
Some examples of annotated common RNA modules are represented. The annotations use the geometric nomenclature where a circle indicates the Watson-Crick sites, the square the Hoogsteen sites, and the triangle the sugar edge sites (filled black, the bases interact in cis; empty symbols, they interact in trans) (Leontis and Westhof 2001). Using this nomenclature, all base–base pairwise interactions present in nucleic acids have been classified in 12 families in which each family is a 4 × 4 matrix of the bases A, G, C, and U. This classification allows to deduce the isotericity matrices that yield all the possible and geometrically equivalent base pairs in a given family. (A) The double loop E motif of bacterial 5S rRNA. The B within the circle indicates a bifurcated pair and a W a water-mediated interaction. The simple motif occurs in the 16S as well as the 23S rRNAs. (B) The S or bulged G motif in which a base triple occurs. This motif is typical of the eukaryotic loop E of rRNA and of the sarcin/ricin loop in bacterial 23S rRNA. The adenines often interact through their Watson-Crick or sugar-edge sites with other nucleotides (Leontis et al. 2002a). (C) The kink-turn motif (Klein et al. 2001). (D) The C-motif (Leontis and Westhof 2003).