Fig. 8.
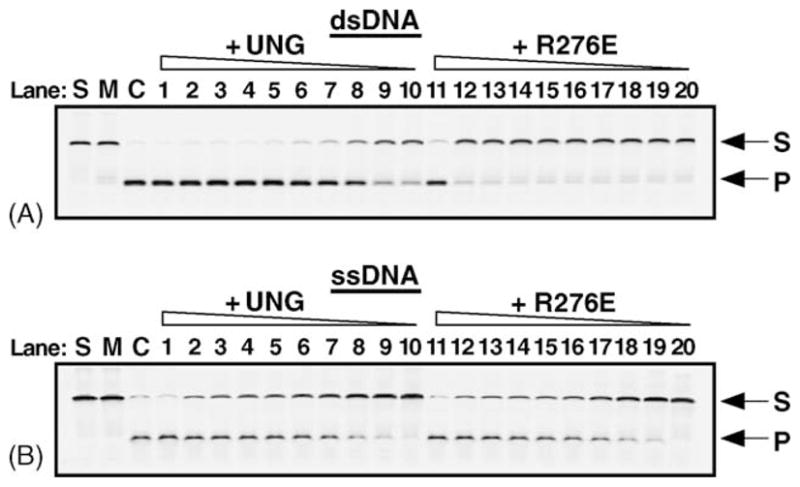
Uracil base excision activity of UNG and R276E on single- and double-stranded uracil-containing DNA. UNG and the R276E mutant enzyme (fraction IV) were assayed for uracil-DNA glycosylase activity using the 5′-end carboxyfluorescein (FAM)-labeled double- and single-stranded uracil-containing oligonucleotide substrates, 5′-FAM-dsU·A-25-mer (panel A) and 5′-FAM-ssU-25-mer (panel B). Control reactions consisted of: S, substrate alone; M, mock reaction mixtures containing substrate in reaction buffer but lacking enzyme; and C, reaction mixtures to which E. coli Ung (88 fmol) was added (lanes S, M, and C, respectively). Reactions contained 1.6 pmol, 80, 40, 30, 20, 17.5, 15, 10, 8.75, and 4.375 fmol of UNG or R276E, as indicated, in lanes 1–10, and 11–20, respectively. Following incubation at 30 °C for 15 min, reaction mixtures that contained duplex 5′-FAM-dsU·A-25-mer (A), were subjected to Endo IV (1 unit) treatment to cleave abasic (AP) sites generated by uracil excision, while the reaction mixtures containing the single-stranded substrate 5′-FAM-ssU-25-mer were subjected to hot alkaline treatment in order to cleave AP sites. Reaction products were analyzed by 12% denaturing polyacrylamide gel electrophoresis and the gels were scanned using a FMBioII fluorescence imaging system. Arrows indicate the locations of the 5′-FAM-ssU-25-mer oligonucleotide substrate (S) and 5′-FAM-10-mer uracil-excision product (P).
