Abstract
Multi-state molecules and multi-component complexes are commonly involved in cellular signaling. Accounting for molecules that have multiple potential states, such as a protein that may be phosphorylated on multiple residues, and molecules that combine to form heterogeneous complexes located among multiple compartments, generates an effect of combinatorial complexity. Models involving relatively few signaling molecules can include thousands of distinct chemical species. Several software tools (StochSim, BioNetGen) are already available to deal with combinatorial complexity. Such tools need information standards if models are to be shared, jointly evaluated and developed. Here we discuss XML conventions that can be adopted for modeling biochemical reaction networks described by user-specified reaction rules. These could form a basis for possible future extensions of the Systems Biology Markup Language (SBML).
Keywords: mathematical model, molecular interaction networks, mashine-readable description, XML, SBML, rule-based, multi-component complexes
I. INTRODUCTION
A. Background
A problem that one confronts when attempting to model a signaling system is combinatorial complexity: the need to consider a very large number of distinct chemical species. Molecules involved in signaling each can be modified in a number of ways, can transition between different functional states, and can combine to form a variety of multi-component species. Typically, a multi-component species consists of several molecules, often proteins or peptides that are associated into a complex. Each protein itself can be viewed as a multi-component species itself by taking into account the multiple functional domains and active sites [1]. Some domains serve as binding sites for bimolecular interactions via recognition of specific regions of partner proteins and other biomolecules. For example, the Src homology 2 (SH2) domain recognizes phosphorylated tyrosines. Protein-protein interactions can be forestalled by modifications of protein domains, such that covalent binding of a phosphate group to a tyrosine residue of a protein substrate (phosphorylation). These modifications can be reversed (e.g., a tyrosine can be dephosphorylated). Protein-protein interactions may be affected by other protein domains, which are not directly participating in binding interactions, such as catalytic domains. Thus, to model protein-protein interactions, we need to identify and describe multiple components within each of the interacting proteins, as well as the full range of species that arise during interactions. The problem of keeping track of all the species and components has been recognized as a serious challenge by many modelers [2–8]. Currently, the problem of generating and analyzing reaction networks while accounting for multi-state multi-component species is addressed by several teams, and a few modeling software packages have been specifically developed to tackle this problem (e.g. StochSim [9], BioNetGen [10], Moleculizer [11]).
B. Combinatorial complexity and SBML
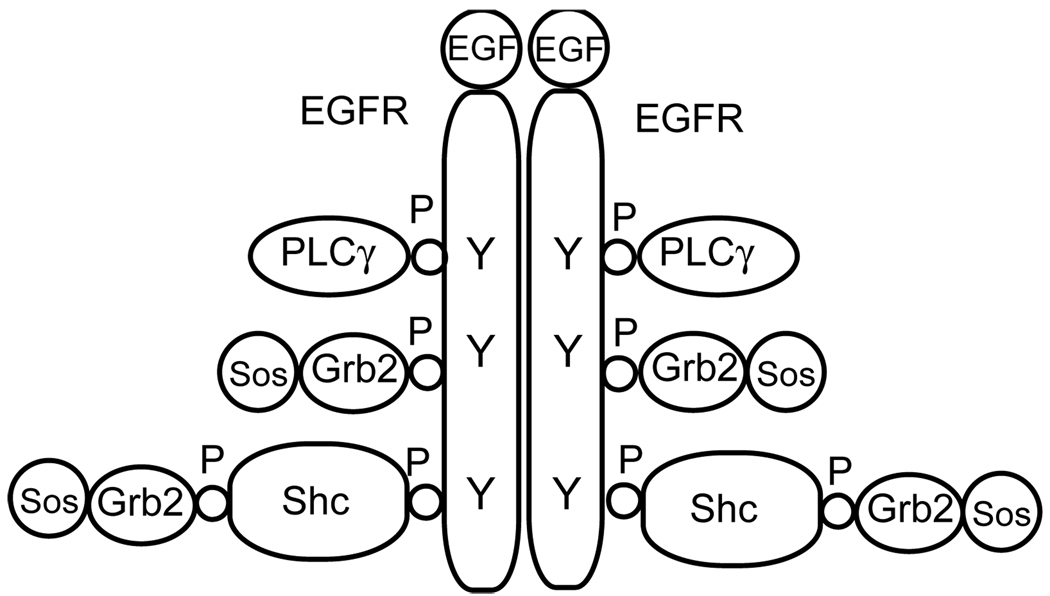
Let us illustrate the problem of combinatorial complexity for the case of epidermal growth factor (EGF) receptor (EGFR) signaling. EGFR is a receptor tyrosine kinase that consists of multiple tyrosines that upon phosphorylation interact with multiple adapter proteins like Shc and Grb. The model [8] for interactions of recruitment of guanine nucleotide exchange factor Sos through EGF-induced formation of EGFR-Grb2-Sos and EGFR-Shc-Grb2-Sos assemblies accounts for 356 distinct chemical species participating in 3749 reactions (see Fig. 1). These species and interactions are specified using a rule-based approach [12, 13] and are generated automatically by a general-purpose software, BioNetGen [10]. A modeler has to use his or her knowledge of the system to provide the following information: (1) molecules to be modeled and their interacting and modification domains, and (2) rules of activities and interactions among domains and molecules.
Figure 1.
Schematic illustration of a protein complex considered in the model of early events in EGFR signaling considered in [8]. All potential pair-wise protein-protein interactions are realized.
This information is then used by to generate: (1) A reaction network, including a set of all chemical species corresponding to specified molecules, and a set of all transitions among these species, with one reaction rate assigned to all reactions among species satisfying specified conditions; (2) Functions of sets of species that correspond to measured quantities (for example, the sum of the concentrations of species with a particular characteristic, e.g. EGFR recruited Sos).
All these outputs can then be written to a file in Systems Biology Markup Language (SBML) Level 2 (L2) format. SBML is an XML-based emerging community standard to encode quantitative models that is being supported by many simulation software tools [14]. Thus, the SBML output can be used to simulate the dynamics of the signaling network. But, a declaration of each individual species and reaction may be not practical for describing of large biological models that account for several thousand chemical species, even in the case of computer software. A BioNetGen software input file describing EGFR receptor signaling system (5 proteins, including a receptor with two tyrosines) consists of 160 lines. A generated model for EGFR consists of 3749 reactions among 356 chemical species. Its representation in SBML L2 format consists of more than 50,000 lines, and takes 12.5 MB of memory – but needs numerical solving of many thousands of coupled ordinary differential equations for dynamic simulations. Moreover, the information about domains and their activities and interactions is not contained in a reaction network written in SBML L2, which provides a flat list of species and interactions only. It is practically impossible to create a human-understandable scheme to visualize such a network, and to provide and verify all necessary input data (such as initial conditions) is difficult at best.
C. Goals of the proposed XML format
We envision a new XML format for describing such complex reaction networks where species and reactions are not explicitly enumerated. Instead, components of bio-molecules (such as tyrosines) and rules of interactions among them are specified. The full range of species and reactions can be generated by appropriate software using these rules. This format provides for compact persistent storage, includes relevant information to enable human understanding and visualization of the signaling networks, and is compatible with the SBML format, which could be extended to facilitate interoperability with many simulator tools.
The desired XML description should be able to:
Store the information about components of multi-molecular complexes, their enzymatic activities (if any), conformational changes, interactions, etc.
Incorporate hierarchical levels of biological information, such as interactions of chemical species, protein domains, individual amino acids, polypeptide chains, sites and domains. A user should be able to specify the level of detail required for a model.
Store the information about experimentally observable features, like all species containing a specific molecule (which could be tracked experimentally by a fluorescent label, for example), or all species with a functional domain in a specific state, etc.
Incorporate species that can be located in different compartments, including species that are located in several compartments simultaneously, such as transmembrane proteins. Define reactions across compartments, including trafficking.
Allow for graphical diagrammatic representation. Required XML standards are currently under discussion by the SBML and SBGN communities.
Be flexible, i.e. a user can change a model by adding or removing certain components without essential changes to the model, e.g. by adding new species or new protein domains and their interactions.
Be expandable, for example, polypeptide chains can be added to the model of protein domain interactions.
II. BIOLOGICAL FEATURES TO BE DESCRIBED
A. Graphical representation
A graph representation for description of biochemical reaction network was introduced by Faeder et al. [15] and formalized by Blinov et al. [16]. The fundamental objects of a biochemical reaction network are "components," collections of which form "molecules," collections of which form "species." Component is the smallest entity that has defined properties. There might be a wide variety of definitions what is a component: it might be a polypeptide chain, a tyrosine, an SH2 domain, a conformational state of a protein, etc. A molecule is defined as a set of components that can be treated as a unit, such as the components of a polypeptide chain or of a multimeric protein.
Graphically, a "species" is represented as a graph. The nodes of the graph are associated with the components, and edges of a graph are associated with the bonds between components. A "molecule" is represented graphically by a box surrounding a set of nodes that represent each component of the molecule. Thus, bonds effectively connect molecules, with each molecule having possibly a large number of bonds, as in Fig. 2a, which illustrates an example of a receptor tyrosine kinase (RTK) dimer stabilized by a bivalent ligand.
Figure 2.
Species graphs of a receptor tyrosine kinase signaling complex. (a) A species graph with chemical bonds between all components declared. (b) A species graph with chemical bonds declared only between molecules. Intramolecular bonds are assumed and not shown. (c) A species graph with labels defining names of components. (d) A species graph with labels defining names and internal states of components, such that PTK is in “A” (active) state, and tyrosines Ya and Yb are either in “P” state (phosphorylated), or “0” state (unphosphorylated).
Edges within molecules may be unaffected by signaling, and thus don't need to be specified, as in Fig. 2b. Thus, only edges that are subject to addition or removal during signaling are declared. Although components are denoted by identical nodes, they all represent different functional domains, thus, they are assigned labels (names), e.g. extra-cellular domain (ECD), SH2 domain, protein tyrosine kinase (PTK), phosphotyrosine binding domain (PTB), tyrosine residue (Ya and Yb) etc, as illustrated in Fig. 2c.
B. Attributes of components
Components may be assigned labels declaring the internal states of the component, as in Fig. 2d. States of components can be introduced for several reasons. In some cases, it is simply convenient to introduce states to define complexes - for example when formed complexes can be indicated by the bound state of the components of a scaffold-like molecule [5]. In other cases, in the absence of 3D structural information, we need states to distinguish the conformations of a molecule. After ECD is bound to a ligand, it can undergo conformational changes and it can be in several modification states, e.g ECD-C. Enzymatic activity of PTK domain can be different and we need to distinguish the inactive and active forms of such a kinase.
The internal state may be omitted if modifications of a domain are unknown, as for the case of the SH2 domains. In some cases, component states are not strictly required but they are biophysically justified and they simplify and/or clarify the representation. For example, consider a phosphorylation-dependent interaction between Y tyrosine of EGFR protein and SH2 domain of Shc protein. The SH2 domain interacts with Y only after Y becomes phosphorylated. It is not physically accurate (and it is ambiguous) to represent this interaction as a linear graph with three nodes (Y, p, and SH2) and two unlabeled edges as follows: Y-p-SH2, as in Fig. 3a. The tyrosine Y and SH2 actually interact, with the interaction being affected by the phosphorylation state of Y. Thus, a more realistic graph looks like that of Fig. 3b. It is more accurate to write Y(p)-SH2, where (p) indicates that the tyrosine is in the phosphorylated state, as in Fig. 3c. One could write Y(u) to indicate the un-phosphorylated state. The introduction of the (p) and (u) states simplifies the representation, because now it is unnecessary to label or interpret the meaning of the edges. With Y-p-SH2, one must understand that the bond between the tyrosine and the phosphate group is a covalent bond and that the bond between the phosphotyrosine and the SH2 domain is a non-covalent bond. One needs to know this because it is impossible for the phosphate group covalently bound to the tyrosine to leave with the SH2 domain in a dissociation reaction. Thus, Y(p)-SH2 is clearer, because one could never make the mistake of allowing the phosphate group of the tyrosine to become associated with the SH2 domain. In this representation, it is clear that the phosphate group is covalently bound to the tyrosine, and not to the SH2 domain.
Figure 3.
Different schemes for declaration of interaction between phosphorylated tyrosine Y and SH2 phosphotyrosine-recognition domain.
C. Bonds
Graph description of multi-component species can be specified with and without explicit declaration of bonds. One can specify bond ends only, which would have an advantage of a shorter form. However, specifying bonds has some advantages. Bonds closely resemble edges introduced for graph XML description like GraphML (http://graphml.graphdrawing.org). In these descriptions edge connects two nodes in the way similar to bond connects two components or species. Thus, no additional processing may be required to represent species as graphs in tools working with graphical XML standards. Moreover, specifying bonds allow for adding additional attributes (e.g. type="covalent").
D. Compartments
SBML L2 defines a Compartment reference requirement for each Species. This does not have implications for the mathematics of the model, and can be ignored by simulators. However, such information is crucial for proper visualization and understanding of reaction networks models, and for analyzing physical constraints and modeling approximations. The optional SpeciesType element allows grouping of like Species with different localizations (in different compartments), but is limiting. Moreover, multi-component species, such as transmembrane receptors, can be located in multiple compartments. Thus, the attribute compartment can be used to track species belonging to the same compartment and to compute observables. Namely, Compartment can be a feature of a component, a molecule or a whole species: a receptor component corresponding to the extracellular portion of a receptor may have a compartment “extracellular”, a receptor component corresponding to tyrosines located in the intracellular portion of the receptor may have compartment “cytosol”, and the receptor as a species have a compartment “membrane”.
We have not attempted to address here another related limitation of SBML L2, namely the lack of localization information for Reaction. If practice will deem it necessary, we will include such information.
E. Group-theoretical operations
The current level of SBML operates on uniquely defined entities. However, when we generalize species and reactions and start working with sets of species, we need to include from MathML group-theoretical operations. Some work may need to be done to map SpeciesTypes to sets. Say, if we want to identify a pattern that selects species with a given components in state 1 or in state 2, we may want to declare
-
<SpeciesPattern id=” State1_or_state2”>
-
<set>
-
<apply>
<union/>
<ci type="set"> A </ci>
<ci type="set"> B </ci>
</apply>
-
</set>
-
</SpeciesPattern>
Here set A is defined as SpeciesPattern with a certain component in state 1, and set B is defined as SpeciesPattern with a certain component in state 2. Similarly, we will need to introduce subsets to define SpeciesPatterns which selects species with components taking values in a certain subset.
F. Open questions: simulation specifications, stochastics
SBML L2 provides description of the reaction system and does not contain simulation specifications, such as the time of simulation, or the type of numerical solver to be used. However, when dealing with rules, these factors can significantly affect the system. Consider, for example, a set of rules that govern formation of polymers, such as actin filaments (Fig. 4). These rules can potentially generate an infinite chain. This can be prevented by simulation directives to truncate chains above a certain length, or by the use of variable kinetic parameters for chain elongation (e.g. rate of elongation becomes zero when the polymer length exceeds a certain value). Another restriction on the length of the polymer is provided by the total number of actin molecules in a cell. These issues are related to stochastic simulation and should be addressed in conjunction with the XML standards for stochastic simulations.
Figure 4.
The simplest rule for actin polymerization: whether there is a barbed end of a polymer, and an actin monomer, they can bind with a certain rate law. This rule can potentially generate an infinite chain. (b) This rule can be expressed by introducing two components of an actin monomer – b (barbed end) and p (pointed end). Unspecified component of the second monomer means that it can be bound to any barbed end of an actin monomer or a polymere.
III. DESIGN OF THE XML REPRESENTATION
Since the ultimate goal is to be able to effectively simulate models of reaction networks that involve multicomponent species and combinatorial complexity, we have built this XML representation on the existing standard of SBML. We will describe two approaches to the encoding of the required XML features.
First, we created a full specification encoded in new classes extending the current SBML schema, as described in Section 4 below. This approach takes advantage of the existing SBML standard to handle all basic entities and operations involving reaction networks, and can be immediately implemented by a simulation software as a proprietary extension. Rules for network generation are defined through new syntactic objects that allow keeping species and reactions in SBML L2 intact. If no changes are required to existing SBML objects, the Annotation element in SBML provides a powerful and flexible way to include additional information encoded by other XML namespaces for proprietary extensions (or any other RDF-type standard or specification). The SBML syntax is intact, and Annotation elements can be ignored. However, a particular software package can use this information to enable additional functionality. For example, we have prototyped enabling the Virtual Cell (VCell, [17–18]) to use this new XML namespace to implement rule-based reaction network generation and visualization as described above.
Second, we developed an alternate specification (described in Section 5 below) which is more intimately linked with the existing SBML standard, but involves changes to the current SBML L2 schema and objects. This could provide the basic framework for an SBML Level 3 (L3) extension (the SBML community has adopted a modular extension mechanism for evolving the SBML language beyond L2). Obviously, the first implementation could be adopted as is in a future L3 extension. However, if most simulation tools that support SBML will eventually support rule-based model descriptions, a tighter integration of the new elements with the core of the SBML language may be desirable. Any such changes to the core would create compatibility requirements for any SBML L3 tool. Therefore, the choices of how to eventually include these features in a future SBML L3 extension will be decided by the members of the global SBML community.
One important choice in both of these approaches is the level and granularity of hierarchical nesting. We can follow the simplest notion that all biological objects are constructed of indivisible objects (molecular species) that can have components and sites (described in section 5.1). The more complex, but possibly more flexible approach would be not to introduce the smallest element, but operate on the abstract level of SpeciesTypes or similar constructs, as described in section 5.2).
IV. THE DECOUPLED XML SPECIFICATION: NEW CLASSES DEFINED AND THEIR BASIC USAGE
This implementation encodes all functionality into new objects that extend existing SBML classes. It can be used as a proprietary extension by simulation software while maintaining full SBML L2 compatibility. The key features of the XML representation of interaction rules and multi-component species are enabled by the new classes Component, ComplexSpecies, SpeciesTemplate, ReactionTemplate. Three new utility classes are also required, ReactionRules, ListOf_ComplexSpecies, and ListOf_ReactionTemplates. In the SBML schema, ComplexSpecies extends Species, whereas the other classes directly extend SBase
A ComplexSpecies is the general representation for a multi-state multi-component entity. It is a container having as possible elements Components (see below; minimum one required), simple Species (the current L2 class; optional), as well as other ComplexSpecies (optional). It thus forms a hierarchy and can be represented as a connected acyclic graph. One could derive all theoretically possible configurations of the entity based on the information in the instance of this class and of its sub-elements. Instances of ComplexSpecies should be defined in an instance of helper class ListOf_ComplexSpecies (instantiated as an element of Model).
A Component is the general representation of a group of a number of mutually exclusive states (minimum 2) of a ComplexSpecies. The state may define the consequence of association/dissociation with a binding partner (another component), in which case it must have exactly 2 values (bound, unbound) and have the attribute isBindingSite set to the value true. The state may define internal condition of the component, in which case it may assume one of user-specified values such as “phosphorylated” or “unphosphorylated”.
A SpeciesTemplate is an arbitrary subset from all the possible configurations encoded by a specific ComplexSpecies (a subgraph). SpeciesTemplates thus reflect modeling intent, defining which types of a ComplexSpecies would actually occur in the model and participate in reactions or other transformations. Instances of SpeciesTemplates are used as elements of ReactionTemplates.
A ReactionTemplate is the general representation for reactions and transformations of multi-state multi-component entities. It is a collection of allowable reactions, encoded using specific SpeciesTemplates instances as reactants and specific ReactionRules for component transformations. There is a single common KineticLaw element. Instances of ReactionTemplate should be defined in an instance of a helper class ListOf_ReactionTemplates (instantiated as an element of Model).
A simulator tool could operate directly using these classes, and graphical representation tools can map this information to a more human-understandable compact form. Conversely, a fairly simple algorithm can be designed to produce a “flattened” L2 compatible SBML document based on the elements in these classes which can be used by other tools. For convenience and to help with backwards compatibility, this algorithm could be implemented in a standalone library or even included in libSBML. See Fig. 5 for examples of ComplexSpecies.
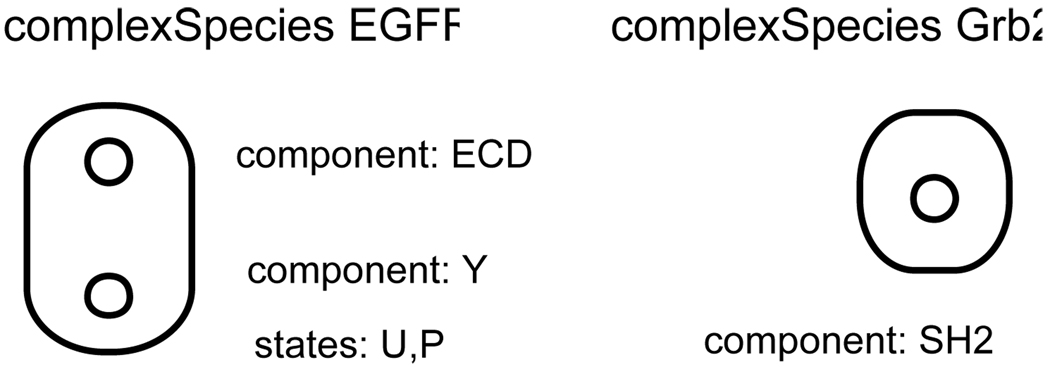
Figure 5.
ComplexSpecies EGFR has two components: ECD and Y, with Y taking states U and P. ComplexSpecies Grb2 has a single component SH2 that can be “bound” or “unbound”, but can not take any internal values.
ReactionTemplate for binding of Grb2 to EGFR will include SpeciesTemplates for reactants:
templ1 (ComplexSpeciesRef EGFR; constraints Y = P, Y = unbound)
templ2 (ComplexSpeciesRef Grb2; constraint SH2 = unbound)
and ReactionRule
(ComplexSpeciesRef EGFR; Y = bound to bond 1)
(ComplexSpeciesRef Grb2; SH2 = bound to bond 1)
Compartment and SpeciesType elements. Since the ComplexSpecies class extends Species, it inherits the required attribute defining the Compartment in which it resides. The optional attribute of SpeciesType (optional logical grouping; ignorable by simulators) could be used as is, or an additional form ComplexSpeciesType could be defined (which would extend SpeciesType). We have used the latter approach, in order to deal with some of the existing limitations of SBML regarding compartments and localization information, which are exacerbated in the case of multi-component complexes.
V. OPTIONS FOR SBML L3 INTEGRATION: NEW AND MODIFIED CLASSES AND BASIC USAGE
As discussed above (Section 3), the decoupled XML specification can be itself the basis for an official SBML language extension for L3. Additionally, we discuss how some of the functionality could be included into the core language by changing existing SBML schema objects. The basic usage of both new classes and modified existing classes is being presented.
A. The simple approach – explicitly defined molecules
The following new classes need to be added:
A ComponentType is as a “minimal” indivisible module of a chemical species and is capable of assuming one of any number of user-defined states. A common use of the component might be to define a prototypical phosphorylation site:
-
<componentType id="p-site" name="phosphosite">
-
<listOfComponentTypeStates>
<componentTypeState value="u"/>
<componentTypeState value="p"/>
</listOfComponentTypeStates>
-
</componentType>
Any physicalEntities (see below) whose definition includes this component will have its state space doubled in that all other states of the species may occur with either the unphosphorylated or phosphorylated component.
A PhysicalEntity is an indivisible molecular entity that is comprised of components. The name PhysicalEntity is chosen to be consistent with the term in BioPAX ontology. PhysicalEntity may take a set of different states. For example, a PhysicalEntity that is comprised of 5 components that define phosphorylation sites can be in 25=32 different forms, representing all the possible phosphoforms of a protein declared within the given model.
The following elements of the SBML L2 specification need to be modified:
A SpeciesTemplate is used as pattern that selects any arbitrary user-specified sets of Species (in SBML L2 it relates Species located in multiple compartments). In particular, any PhysicalEntity can be declared as SpeciesTemplate. For the considered example of a protein with 5 tyrosines, SpeciesTemplate would select 32 individual Species. SpeciesTemplate can include several physical entities in specific states, e.g. a complex of two proteins connected via association of SH2 domain of one with the phosphorylated residue of the second. In the last example, the configuration space of this SpeciesTemplate includes all modifications of both proteins that do not break the bond between SH2 and phosphotyrosine. In graph-theoretical language, SpeciesTemplate is a connected graph with vertices that may have different states. A very important feature of the SpeciesTemplate is that it can be “closed” or “open”. A “closed” SpeciesTemplate has all components being unbound, meaning that no other physicalEntities except specified in SpeciesTemplate declaration can be selected. An “open” SpeciesTemplate can select Species that include other physicalEntities (Fig 6b).
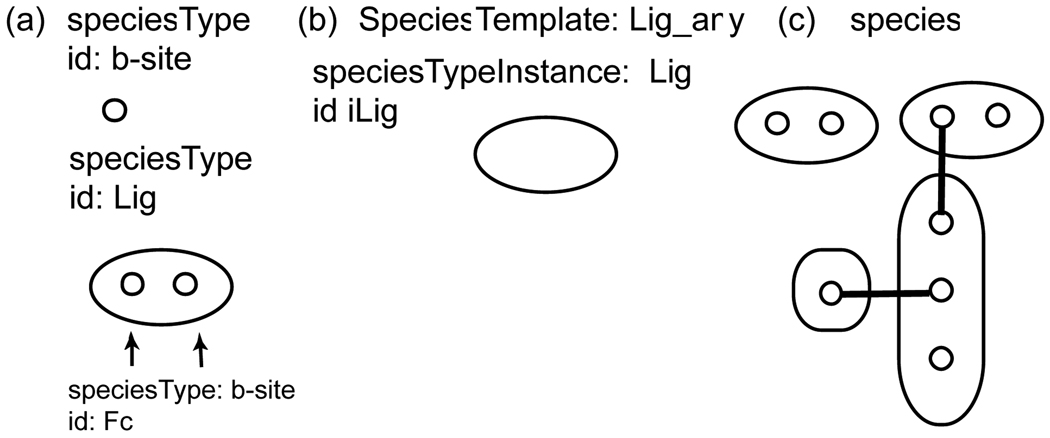
Figure 6.
Schematic representation of elements of SBML extension. (a) physicalEntities are constructed of components b-site (“binding site”). (b) “open” SpeciesTemplate that selects any Species that contain a ligand. (c) Some Species selected by this SpeciesTemplate: a ligand and a ligand in a protein complex.
Species are uniquely defined entities. Species have the same use as in SBML L2: specify initial concentrations and be referenced as reactants or products in reactions. The new feature is that each Species has an internal structure consisting of physicalEntities connected through bonds between components. Each Species may now belong to more than one SpeciesTemplate, e.g. a Species ligand-receptor complex can belong to a SpeciesTemplate ligand and to SpeciesTemplate receptor, if both SpeciesTemplates are open.
Reactions now incorporate reaction rules that operate on Species selected by SpeciesTemplates, if SpeciesReference is replaced with SpeciesTemplateReference. If so, the reaction effectively becomes a reaction rule (not in the SBML sense of Rule, but for generating multiple reactions), although no special tag for reaction rule is introduced. Depending on whether reactants or products are uniquely defined Species, it can be a regular reaction or a reaction-generating rule.
B. The full hierarchical case
One may introduce the fully hierarchical structure of XML data. In this approach, a SpeciesTypes that can be constructed of other SpeciesTypes in the way physicalEntities are constructed of components. The simplest SpeciesType would be a component (such as a phosphorylation site) that may have different states (such as phosphorylated and un-phosphorylated forms). SpeciesType can also be further constructed from other SpeciesTypes connected by bonds, like a graph is constructed of vertices connected by edges. Each Species is represented as a graph with each vertex (SpeciesType) being fully defined (instantiating a single state out of the potential set of states declared in the SpeciesType for this vertex). Edges of this graph are chemical bonds between SpeciesTypes.
In the fully hierarchical case, SpeciesType plays a role of a component or a physicalEntity. We also need to introduce SpeciesTemplate that is used to select any arbitrary user-specified sets of Species. Examples of SpeciesTemplates include: (i) a SpeciesType in a specific state, e.g. if the SpeciesType represents a phosphotyrosine, then having it phosphorylated will select all Species that have a given phosphosite in a phosphorylated state; (ii) a SpeciesType with all but a single embedded SpeciesType having specified states, e.g. a SpeciesType representing a protein with all but one of its tyrosines being phosphorylated; this SpeciesTemplate selects only as many Species as there are different states in the SpeciesType declaration for this phosphosite, e.g. two Species with a given tyrosine being phosphorylated or not.
We can define the different hierarchical levels of SpeciesTypes using controlled vocabularies (CV). CVs can be used to define the names of intermediate elements, which are embedded one into another and can be represented in many different ways:
molecules ← components
proteins ← domains ← components
proteins ← domains ← peptides ← components
proteins ← domains ← peptides ← amino acids
A Reaction may operate on the set of Species selected by a SpeciesTemplates and effectively becomes a reaction-generating rule, if SpeciesReference is replaced with SpeciesTemplateReference.
VI. CONCLUSION
A growing number of standards have been recently developed to facilitate the exchange of biological models between different resources and software tools. However, different types of information use different exchange formats. Pathway data are often encoded using BioPAX ontology whereas simulation-ready quantitative models are typically exchanged in SBML or CellML format, and there are recent efforts to standardize visualization of reaction networks in the form of SBGN. However, no general standard exists with regard to models created and described use molecular interaction rules. This is the only viable approach to deal with the combinatorial explosion of reaction networks involving multi-molecular complexes and molecules with many functional domains and states. Several software tools currently exist that use this approach, but they all use proprietary mechanisms to encode the models. If these models are translated into an explicit reaction network, they can be shared with other tools using the SBML format. However, this does not supplant exchange of the actual models, as essential features that were used to generate the reaction networks (e.g. components of macromolecules, binding rules, etc.) are lost. We described here an XML format that could form the basis for a standard to exchange rule-based models. It is built around three existing technologies: (i) the graph-theoretical description developed by the BioNetGen group, (ii) the functional relationship concepts encoded in the BioPAX ontology, and (iii) the current SBML Level 2 Revision 3 standard. Ideally, this can evolve into a community standard as an SBML Level 3 extension. As described in the text, this could be a decoupled extension, or, for tighter integration, it could involve changes to some of the core SBML classes. The implementation-related decisions were mostly based on the practical needs of two tools: BioNetGen (a software for rule-based modeling of reaction networks originally developed to operate in a single compartment) and the VCell modeling framework (that supports both compartmental and spatial modeling). Recently we integrated BioNetGen into a VCell application (http://vcell.org/bionetgen); however, the integration of these tools can not be complete without a standard that incorporates both rule-based and compartmental features, something we tried cover briefly in this manuscript. This is just one example on how multiple standards should be developed together. Extensive community efforts should eventually lead to an intelligent exchange of biological information on multiple levels: data, models and visualization.
Figure 7.
Schematic representation of elements of proposed SBML extension. (a) SpeciesTypes can be constructed of another SpeciesTypes, as SpeciesType ligand has two SpeciesTypeInstances b-site. (b) “open” SpeciesTemplate that selects any Species that contain a ligand. (c) Some Species selected by this SpeciesTemplate: a ligand and a ligand in a protein complex.
ACKNOWLEDGMENTS
We would like to thank James R. Faeder (Pittsburgh University), William S. Hlavacek (Los Alamos National Laboratory), Michael Hucka (Caltech), Oliver Ruebenacker, and James C. Schaff (University of Connecticut Health Center) for many ideas and helpful discussions regarding this project. The project was supported in part by NIH R01 GM076570 grant (MLB) and NIH U54 RR022232 grant (IIM).
Contributor Information
Michael L. Blinov, Email: blinov@uchc.edu.
Ion I. Moraru, Email: moraru@panda.uchc.edu.
REFERENCES
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Arkin A. Synthetic cell biology. Curr Opinion Biotechnol. 2001;12:638–644. doi: 10.1016/s0958-1669(01)00273-7. [DOI] [PubMed] [Google Scholar]
- 3.Endy D, Brent R. Modeling cellular behavior. Nature. 2001;409:391–395. doi: 10.1038/35053181. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein B, Faeder JR, Hlavacek WS, Blinov ML, Redondo A, Wofsy C. Modeling the early signaling events mediated by FceRI. Mol Immunol. 2002;38:1213–1219. doi: 10.1016/s0161-5890(02)00066-4. [DOI] [PubMed] [Google Scholar]
- 5.Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B B. Investigation of early events in Fc epsilon RI-mediated signaling using a detailed mathematical model. J Immunol. 2002;170:3769–3781. doi: 10.4049/jimmunol.170.7.3769. [DOI] [PubMed] [Google Scholar]
- 6.Hlavacek WS, Faeder JR, Blinov ML, Perelson AS, Goldstein B. The complexity of complexes in signal transduction. Biotechnol Bioeng. 2003;84:783–794. doi: 10.1002/bit.10842. [DOI] [PubMed] [Google Scholar]
- 7.Bray D. Molecular prodigality. Science. 2003;299:1189–1190. doi: 10.1126/science.1080010. [DOI] [PubMed] [Google Scholar]
- 8.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. Biosystems. 2006;83:136–151. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Le Novere N, Shimizu TS. STOCHSIM: modelling of stochastic biomolecular processes. Bioinformatics. 2001;17:575–576. doi: 10.1093/bioinformatics/17.6.575. [DOI] [PubMed] [Google Scholar]
- 10.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioNetGen: software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 11.Lok L, Brent R. Automatic generation of cellular reaction networks with Moleculizer 1.0. Nat Biotechnol. 2005;23:131–136. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- 12.Faeder JR, Blinov ML, Goldstein B, Hlavacek WS WS. Rule-based modeling of biochemical networks. Complexity. 2005;10:22–41. [Google Scholar]
- 13.Hlavacek WS, Faeder JR, Blinov ML, Posner RG, Hucka M, Fontana W. Rules for modeling signal-transduction systems. Sci STKE. 2006;344:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 14.Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 15.Faeder JR, Blinov ML, Hlavacek WS. Graphical rule-based representation of signal-transduction networks. In: Liebrock LM, editor. Proc. 2005 ACM Symp. Appl. Computing; New York, NY. ACM Press; pp. 133–140. [Google Scholar]
- 16.Blinov ML, Yang J, Faeder JR, Hlavacek WS WS. Graph theory for rule-based modeling of biochemical networks. Transact. Computat. Syst. Biol. VII in the series Lect. Notes Comput. Sci. 2006;4230:89–106. [Google Scholar]
- 17.Loew LM, Schaff JC. The Virtual Cell: A software environment for computational cell biology. Trends in Biotechnology. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 18.Moraru II, Schaff JC, Loew LM. Think simulation – think experiment: the Virtual Cell paradigm. In: Perrone LF, Wieland FP, Liu J, Lawson BG, Nicol DM, Fujimoto RM, editors. Proc. 2006 Winter Sim. Conf; Institute of Electrical and Electronics Engineers, Inc.; Piscataway, NJ. 2006. pp. 1713–1719. [Google Scholar]