Figure 1. Allele-specific chromatin contacts at an imprinted locus.
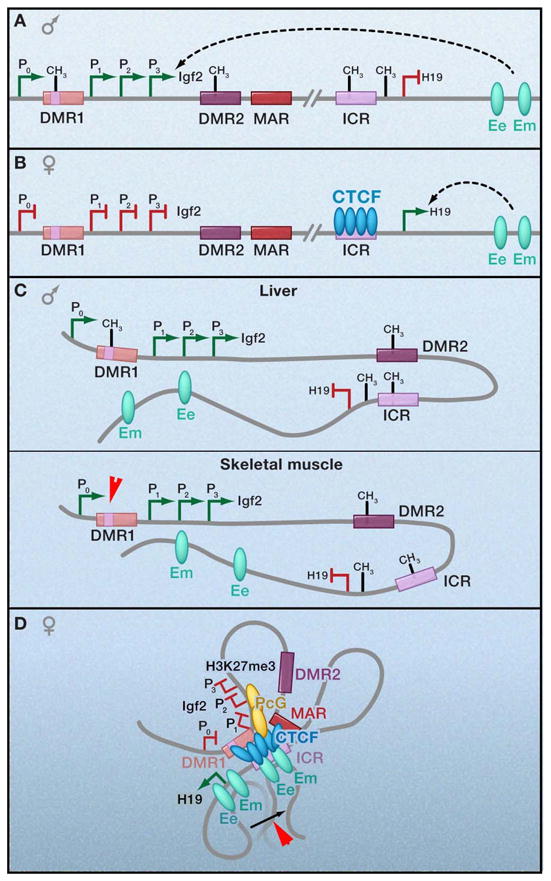
(a,b) Linear depiction of the mouse H19/Igf2 locus. The maternally expressed non-coding H19 gene is located approximately 90kb downstream from the gene encoding Insulin-like growth-factor 2 (Igf2) that is expressed exclusively from the paternal allele. The imprinting control region (ICR) ~2kb upstream of H19 contains four CTCF binding sites and is essential for regulation of the entire locus. Differentially methylated regions (DMRs), such as DMR1 upstream of Igf2 promoters (P1, 2, 3) and DMR2 within Igf2 exon 6, act in concert to regulate reciprocal, allele-specific expression patterns from a shared set of downstream enhancers at 8kb (Ee: endodermal tissue enhancer) and 25kb (Em: mesodermal tissue enhancer) downstream of the H19 gene. –CH3, DNA methylation. Green ovals, enhancers. (c,d) Schematic 3-D models illustrating allele-specific patterns of CTCF binding, DNA methylation, and chromatin looping. Although loops are illustrated here via CTCF multimerization, it is not yet clear if these long-range interactions can be attributed to CTCF binding to all sites and subsequent dimerization or if CTCF detection via chromatin immunoprecipitation is due to indirect interactions via looping.
