Figure 4. Potential classes of CTCF-mediated contacts.
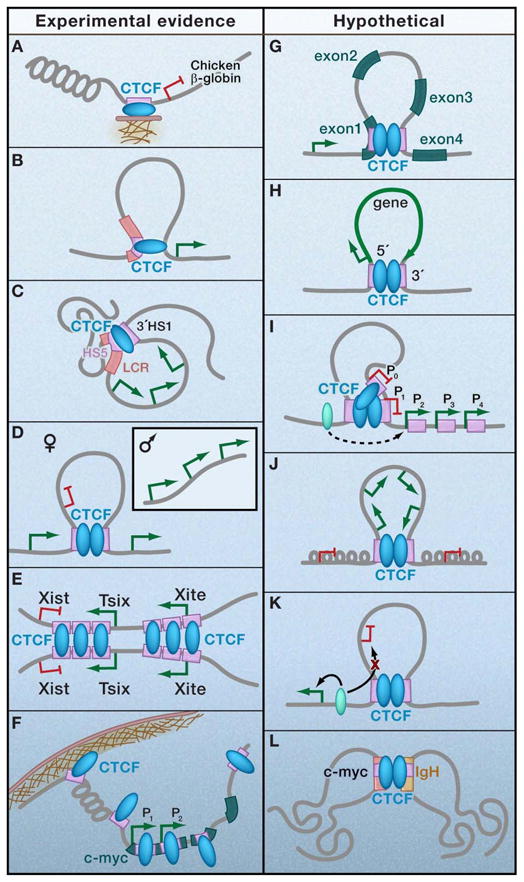
Experimental evidence for certain sub-classes of CTCF loops exists (a–f), whereas others can be hypothesized based on genome-wide distribution patterns (g–l). (a) Anchoring via direct attachment to subnuclear structures such as the nucleolus and/or nuclear matrix; (b) Transcriptional regulation via contact between intergenic locus control region and promoter-proximal regulatory element; (c) Active chromatin hub around multiple co-regulated genes via contact between multiple distal CTCF binding sites; (d) Mono-allelic gene expression via allele-specific contacts between multiple imprinted regulatory elements; (e) X-chromosome inactivation or mono-allelic gene expression via interchromosomal contacts between regulatory elements in trans; (f) Global nuclear organization via demarcation of lamina-associated domains (LADs; (g) RNA polymerase II pausing and/or termination via intragenic contacts between introns and exons; (h) RNA processing or transcriptional re-initiation via a single gene 5′-3′ loop; (i) Alternative promoter selection via contact between two insulator elements demarcating transitions in chromatin structure; (j) Boundary/barrier loops to demarcate independently regulated chromatin domains containing a co-regulated gene-dense cluster via contact between two insulator elements; (k) Enhancer blocking loops that topologically separate inappropriate enhancer-promoter interactions via contact between two insulator elements; (l) Interchromosomal translocations via contacts between two regulatory elements in trans.
