Figure 1. LPS-induced chromatin reorganization within −3 kb cis-element of the lysozyme gene.
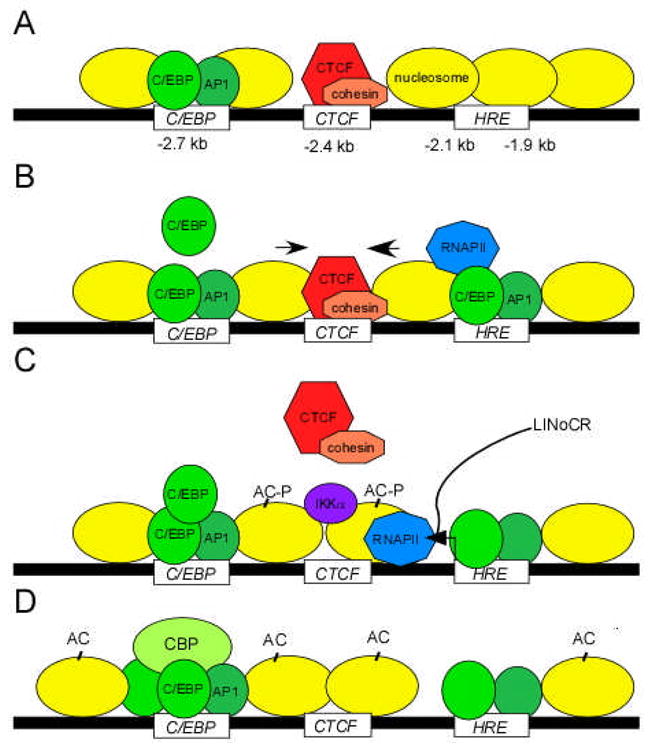
(A) In unstimulated monocytes, CTCF and cohesin form an insulator complex at the −2.4 kb silencer element. (B) Short LPS stimulation induces binding of C/EBPβ, AP1 and RNAPII to the exposed HRE, and the displacement of nucleosomes towards the CTCF occupancy site (black arrows). (C) At the one hour time-point, LINoCR transcription, IKKα recruitment, specific H3 phosphoacetylation (AC-P), and repositioning of a nucleosome over the CTCF site culminate in the eviction of the CTCF/cohesin complex. (D) Prolonged LPS stimulation prevents binding of CTCF/cohesin due to re-positioning of the nucleosome over its site. Recruitment of additional C/EBP and CBP to the −2.7 kb enhancer increases H3 acetylation (-AC) and maintains the lysozyme gene in an active state.
