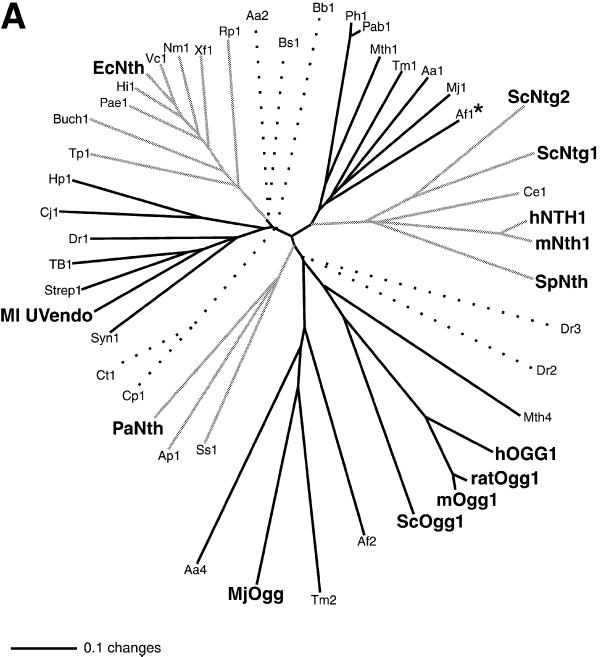
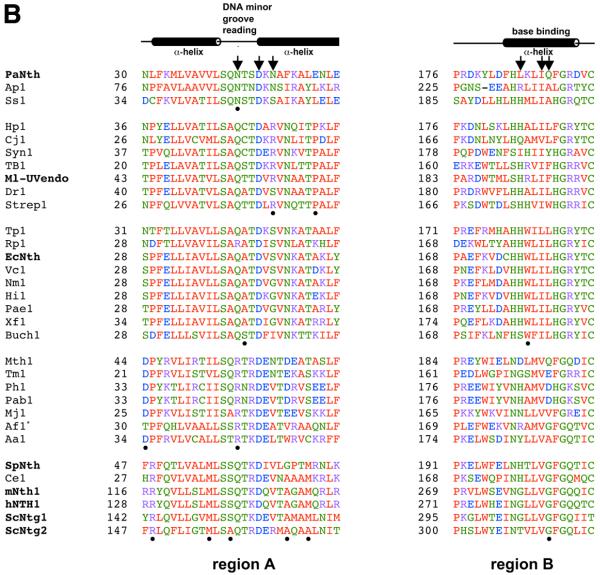
Figure 10.
(A) Phylogenetic analysis of members of the DNA glycosylase superfamily that have a lysine residue in the HhH motif corresponding to Lys120 in EcNth (protein code and GenBank accession number in parentheses). Aeropyrum pernix (Ap1, BAA79061), Aquifex aeolicus (Aa1 AAC06594; Aa2, AAC06742; Aa4, AAC06576), A.fulgidus (Af1, AAB89556; Af2, AAB90876), Bacillus subtilis (Bs1, P39788), Borrelia burgdorferi (Bb1, AAC67089), Buchnera sp. APS (Buch1, BAB12837), Caenorhabditis elegans (Ce1, P54137), Campylobacter jejuni (Cj1, CAB75231), Chlamydia pneumoniae (Cp1, AAD18975), Chlamydia trachomatis (Ct1, AAC68292), Deinococcus radiodurans (Dr1, B75537; Dr2, D75275; Dr3 C75459) E.coli (EcNth, P20625), Haemophilus influenzae (Hi1, P44319), Helicobacter pylori (Hp1, AAD07651), Homo sapiens (hNTH1, AAC34209; hOGG1, CAA10351), Methanobacterium thermoautotrophicum (Mth1, AAB85267; Mth4, AAB85820), Methanococcus jannaschii (Mj1, AAB98606; MjOgg, Q58134), M.luteus (Ml UVendo, P46303), Mus musculus (mNth1, BAA22080; mOgg1, AAB61289), Mycobacterium tuberculosis (TB1,CAA17996), Neisseria meningitidis MC58 (Nm1, AAF40962), Pseudomonas aeruginosa (Pae1, AAG06883), Pyrobaculum aerophilum (PaNth, AAF37269), Pyrococcus abyssi (Pab1, A75109), Pyrococcus horikoshii (Ph1, BBA30606), Rattus norvegicus (ratOgg1, AAC77525), Rickettsia prowazekii (Rp1, CAA72458), Saccharomyces cerevisiae (ScNtg1, AAC04942; ScNtg2, CAA99045; ScOgg1, AAC49312), Schizosaccharomyces pombe (SpNth, CAA91893), Streptomyces coelicolor (Strep1, T36554), Sulfolobus solfataricus (Ss1, CAA69576), Synechocystis sp. (Syn1, P73715), Thermotoga maritima (Tm1, Q9WYK0; Tm2, Q9X2E1), Treponema pallidum (Tp1, AAC65744), Vibrio cholerae (Vc1, AAF94172), Xylella fastidiosa (Xf1, AAF83457). The bar scale stands for number of substitutions per site. Bold labels designate proteins with biochemically determined functions. The asterisk marks the endonuclease III homolog from A.fulgidus (Af1, AAB89556) which has been studied only by NMR structure (48). To help differentiate cluster groups, we use lines of different shades; dotted lines indicate homologs, which apparently do not fall into any of these existing groups. (B) Sequence alignment of regions A and B for phylogenetic groups of endonuclease III/UV endonuclease homologs using the program ClustalW Alignments in Color (68; http://www2.ebi.ac.uk/clustalW). Putative secondary structure assignments, according to the crystal structure of E.coli endonuclease III (37), are listed at the top. The program colors amino acid residues as follows: red, AVFPMILW (small and hydrophobic); blue, DE (acidic); magenta, RK (basic); green, STYHCNGQ (hydroxyl and amine). The dots mark amino acid residues occurring >80% of the time within a single group and outside the group only once or not at all. The arrows mark amino acid residues corresponding to residues in E.coli AlkA and human hOGG1, which are involved in protein–DNA interactions (41,42). Bold labels designate proteins with biochemically determined functions. For positions of regions A and B, with respect to the whole protein sequence, see Figure 1.