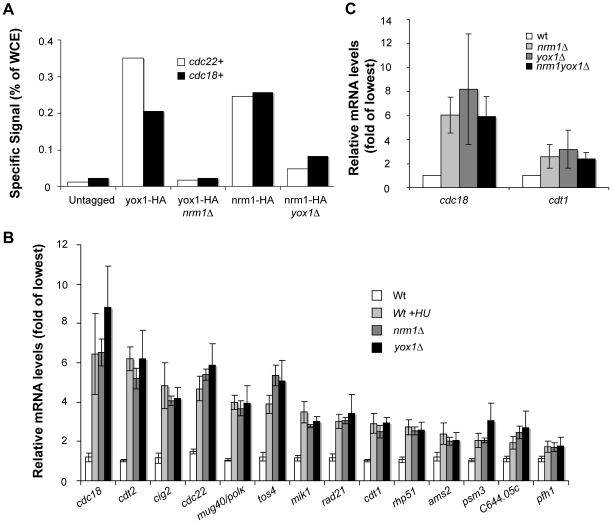
Figure 2. Yox1p and Nrm1 require each other to bind and repress MBF targets.
(A) Chromatin-immune precipitated (ChIP) cdc22 and cdc18 promoter fragments pulled down by HA tagged Nrm1 and Yox1 in wild type, and yox1Δ and nrm1Δ cells, respectively, were quantified by qPCR. Enrichment is shown as percentage of WCE signal. Untagged cells were included as negative control and data shown are representative of multiple independent experiments (see Figure S1 for a biological repeat experiment). (B) Relative mRNA levels obtained by RT-qPCR for 14 MBF-dependent transcripts in untreated and HU treated wild type cells and nrm1Δ and yox1Δ cells. Transcript levels are shown as fold induction of transcript levels detected in wild type untreated cells. Bars represent the average value, and error bars represent their SD, obtained by qPCR of triplicate biological samples. (C) RT-PCR analysis of the relative levels of cdc18+ and cdt1+ transcripts in wild type, nrm1Δ, yox1Δ and nrm1Δyox1Δ cells in untreated conditions and as percentage of maximal levels (100%). Bars represent the average value, and error bars represent their SD, obtained by qPCR of triplicate biological samples.