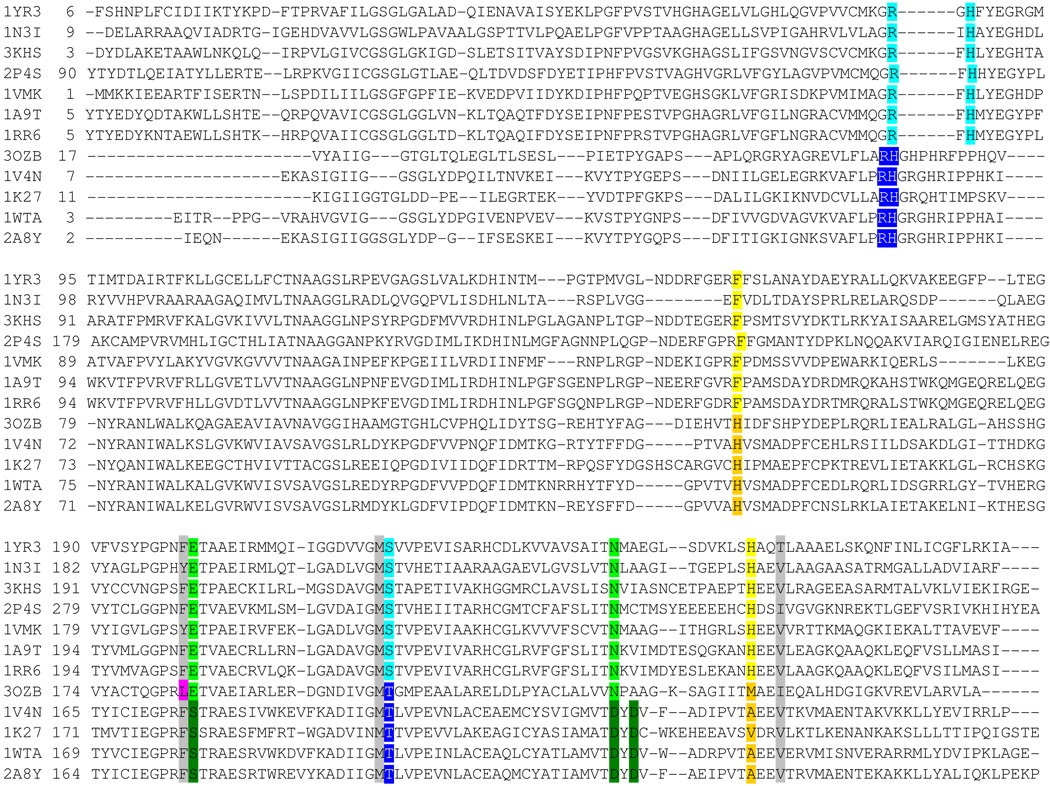
Figure 4.
Structure-based sequence alignment. Protein are listed by PDB ID. The top 7 sequences are PNPs, the bottom 4 sequences are MTAPs and PaMTIP is 3OZB. The conserved residues in phosphate binding region of PNPs and MTAPs are shaded in light blue and dark blue, respectively. The conserved residues in the purine binding region of PNPs and MTAPs are shaded in light green and dark green, respectively. The conserved Phe and His in ribose-binding region of PNPs are shaded in yellow. The conserved His and small hydrophobic amino acid in the (methylthio)ribose-binding region of MTAPs are shaded in orange. The active site residues conserved in all species are shaded in grey. The Leu of PaMTIP is not conserved to either MTAPs or PNPs and is shaded in pink. The PDB IDs are as follow: 1YR3, E. coli PNP II; 1N3I, Mycobacterium tuberculosis PNP; 3KHS, Grouper Iridoviurs PNP; 2P4S, Anopheles Gambia PNP; 1VMK, Thermotoga maritime PNP; 1A9T, bovine PNP; 1RR6, human PNP; 3OZB, PaMTIP; 1V4N, Sulfolobus tokodaii MTAP; 1K27, human MTAP; 1WTA, Aeropyrum pernix K1 MTAP; 2A8Y, Sulfolobus solfataricus MTAP.