Abstract
Protein N-glycosylation requires the presence of asparagine (N) in the consensus tri-peptide NXS/T (where X is any amino acid, S is serine and T is threonine). Several factors affect the glycosylation potential of NXS/T sequons and one such factor is the type of amino acid at position X. While proline was shown to negatively affect N-glycosylation, the nature of other amino acids at this position is not clear. Using Markov chain analysis of tri-peptide NXS/T from viral, archaeal and eukaryotic proteins as well as experimentally confirmed N-glycosylated sequons from eukaryotic proteins, we show here that the occurrence of most sequon types differ significantly from the expected probability. Sequon types with F, G, I, S, T and V amino acids are consistently preferred while those with P and charged amino acids are under-represented in all four groups. Further, proteins contained far fewer number of possible sequon types (maximum 20 types for NXS or NXT taken separately) for any given number of sequons, which may be explained based on random sampling. Consistent with the present finding, majority of the over-represented sequons found in two important viral envelope glycoproteins (hemagglutinin of influenza A H3N2 and glycoprotein120 of HIV-1) are indeed preferred sequon types, which may provide a selective advantage. Accordingly, although there seems to be some preference for sequons, this preference may not be unique to N-glycosylation.
Keywords: HIV, Influenza, N-glycoproteins, Probability, Sequons
Background
Most proteins require some kind of co/post-translational structural modifications for proper functioning. Asparagine (N) linked glycosylation is an important co-translational modification which affects a number of properties of proteins such as folding, half-life, transport and immunogenicity [1, 2]. N-glycosylation requires the presence of asparagine in the consensus tri-peptide NXS/T (where N is asparagine, S is serine, T is threonine and X is any amino acid) [1– 3]. Although NXS/T tri- peptides (also called sequons) occur frequently in many proteins, N- glycosylation occurs only on a subset of sequons found in membrane- bound/secretory proteins which are exposed to the enzyme oligosaccharyltransferase in the lumen of the endoplasmic reticulum [3– 6].
While this indicates that NXS/T sequons are essential for N-glycosylation, the presence of a sequon by itself is not sufficient. That is, there could be many other local and global cues required to determine whether and if, to what extent a sequon is N-glycosylated. These include the position/neighborhood of sequons in the protein, primary sequence and secondary/tertiary structure of the protein, and whether it is destined to cell-membrane/secretion [ 4–6]. Among many factors that are known to affect the N-glycosylation potential of sequons, the nature of NXS/T tri- peptide itself is the important one. For example, although eukaryotic proteins in general contain more number of NXS sequons as against NXT, N-glycosylation occurs less frequently at NXS sequons compared to NXT [3, 5,6].
Similarly, different NXS/T sequon types (based on the type of amino acid at position X) may show a higher/lower potential for glycosylation. Based on a set of 417 N-glycosylated sequons from around 50 eukaryotic proteins, Gavel and von Heijne [3] concluded that the presence of proline in sequons greatly reduces the likelihood of N-glycosylation. This has been attributed to the rigid/unfavorable conformation of proline in the sequon [3,5, 6].Possibly due to the small sample size, previous work [3] did not find any further sequon features that could have a significant affect on the glycosylation status of the sequons. However, apart from proline containing sequons, other types of sequons may have a differential influence on the potential for glycosylation [7, 8]. For example, in two recent studies using slightly larger data sets, it has been shown that sequons containing G, T, W and C are over-represented in glycoproteins, and E, R, K and M are under-represented [5, 6]. Interestingly, it has been also shown, for example in rabies virus glycoprotein, that NXS sequons containing W, D, E and L amino acids were poorly glycosylated in vitro compared to their NXT counterparts [7]. These observations indicate that different types of sequons might have differential potential for glycosylation and, the NXS and NXT sequons behave differently. However, results from these works lacked overview/scope due to the small sample size or case study.
Taking these prior details into consideration, we asked the following questions. Do N-glycoproteins and proteins in general (which contain NXS/T sequons) have a preference for some sequon types over others? What is the average number of sequon types for a given number of sequons in proteins? And finally, whether viral envelope glycoproteins exploit the advantage of using preferred sequons?
Our objective was not to discriminate glycosylated sequons form non- glycosylated ones, but to explore the differential glycosylation potential of sequon types. Accordingly, we sought to answer the afore mentioned questions using a systematic analysis of sequons collected from a large set of experimentally confirmed eukaryotic N-glycoproteins and NXS/T tri peptides fromeukaryotic, viral and archaeal proteins (although it is known that the N-glycosylation occurs in some bacteria, this group was not included in this study)[9– 12]. In addition, we analyzed two specific viral envelope glycoproteins (hemagglutinin of influenza A H3N2 andglycoprotein-120 of HIV-1) with regard to their preference for types of sequons [[10], [13–15]].
Methodology
Sequence/sequon acquisition
Non-redundant and reviewed sets of viral, archaeal and eukaryotic protein sequences were downloaded from the Swiss-Prot part of the Uniprot database [16]. Sequences shorter than 100 residues and those with NXS/T sequons were removed. Proteins with more than 70% identity with other proteins were removed by using sequence block alignment (41 residue window centered at sequon asparagine) to reduce the effect of large protein families/homologous proteins [3, 11]. Final sets consisted of 23596 viral (6313 proteins), 12365 archaeal (6476 proteins) and 264251 eukaryotic (73425 proteins) NXS/T tri-peptide sequences.
The influenza A H3N2 hemagglutinin (HA) sequences (1319) were downloaded from the Influenza Virus Resource at National Centre for Biotechnology Information [17] and the HIV-1 glycoprotein-120 (gp120) sequences (11333) were downloaded from HIV database at Los Alamos National Laboratory [18, 20].
The resource description framework (RDF) files of viral, archaeal and eukaryotic proteins downloaded from Swiss-Prot were parsed and N- glycosylation sites containing a reference tag (citation indicating the experimental evidence) for N-linked glycans were gathered [3, 4]. In total, 4035 experimentally confirmed N-glycosylated sites were collected from 1793 eukaryotic proteins (157 species). Due to the small sample size, viral and archaeal sites (70 and 24 respectively) were not used. Proteins with more than 70% identity with other proteins were discarded and the final set consisted of 3324 experimentally confirmed N-glycosylated sequons from 1553 eukaryotic proteins.
Number and types of sequins
For each protein, the number of sequons was counted and the sequon type was identified based on the central amino acid X in NXS or NXT. Proportion of each sequon type was calculated from pooled sequons for each (viral, archaeal or eukaryotic) group. The amino acid proportions were calculated from the pooled amino acid frequencies for each group. The expected probabilities of sequon types are equal to the proportions of individual types of amino acids according to zero order Markov chain [19, 20].
For HA and gp120 sequences, the actual sequon density was considered the number of sequons per 100 amino acid residues. The expected sequon density was calculated from the second order Markov chain [19, 20].
Data analyses
All the sequence analyses and data handling were done using programs written either in Python (ver. 2.6) or in C programming language. A Microsoft Excel 2003/2007 spreadsheet was used to visualize the data and SigmaPlot (ver. 11, Systat Software Inc, CA, USA) was used for the contour maps. A Z-test for two proportions was performed between expected versus observed sequon types and was declared significant at p ≫ 0.05. For the simulation of probability sampling, sequons were drawn randomly from a set of 20 possible sequon types (proportional to amino acid probability) and means of 100000 samplings were given for each data point.
Discussion
Preference for sequon types
The proportion of different types of sequons (NXS and NXT separately) in viral, archaeal and eukaryotic proteins are presented in Table 1. Here, the expected proportion of sequon types is equal to the proportion of different amino acids in a protein according to zero order Markov chain. That is, given 20 types of amino acids, a maximum of 20 sequon types (NXS and NXT separately) may be expected in a protein [19,20]. Table 1 also shows the observed proportion of sequon types for experimentally confirmed N- glycosylated sites in eukaryotic proteins. It can be seen that the observedproportions of proline containing sequons (NPS and NPT) are greatly under-represented (10 NPS/T out of 3324 glycosylated sequons) compared to the expected proportions. This is in accordance with the earlier study of 417 N-glycosylated sequons where Gavel and von Heijne [3] showed that the presence of proline in sequons strongly reduced the likelihood of N-glycosylation. This has been attributed to the unfavorable conformation of proline containing sequons. Perhaps due to small sample size, the previous study [3] did not find any further local sequence feature differences among glycosylated versus non-glycosylated sequons. Our aim was not to differentiate glycosylated sequons from non-glycosylated ones, but to differentiate sequon types. Accordingly we found that the occurrence of most sequon types differed significantly (p ≫ 0.05, Z-test for two proportions) from the expected probability [3, 8]. Sequon types with G and A (small amino acids), S and T (hydroxyl amino acids), C, I, V (hydrophobic amino acids), and F and Y amino acids were preferred, whereas sequons containing P, Q and charged amino acids (D, E, R and K) were under-represented [5,7]. Interestingly, this preference was not unique to N-glycoproteins since nearly a similar pattern of preference for NXS/T tri-peptide sequence types was found in general eukaryotic proteins. Such a distinction can even be made for NXS/T tri-peptide sequence types in viral and archaeal proteins [9]. This could be due to the positive selection of preferred sequons during the evolutionary time [11]. It should be noted that in all four groups there is slight variation in the preferences, and disparity among NXS and NXT sequons [7]. Thus, the sequon preference in N-glycosylation appears to be in line with the overall sequons in proteins.
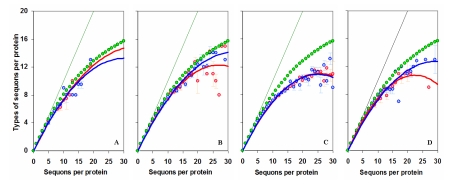
Proteins show fewer sequon types
If proteins have preference for some sequon types over others, then it may be expected that such proteins must show a reduced likelihood to represent all possible sequon types for any number of sequons. For example, if there are 20 sequons (NXS and NXT separately) in a protein, then that particular protein can have all 20 possible sequon types. However, as seen in Figure 1 A to D, proteins show far fewer number of possible sequon types. This trend is similar in eukaryotic N-glycoproteins and other proteins containing NXS/T tri-peptide sequences. It is interesting to note that the observed pattern may be the result of bias towards some sequon types [3–8], but it can also be explained on the basis of a random process of probability sampling. That is, given n types of objects, a probability sampling of n objects yields lower than the possible n types of objects. Therefore, the observed trend in the number of sequon types versus number of sequons in proteins is likely to be the combined result of some selective process for specific sequons [5–7] and a random phenomenon [19,20]. This is the first study to shown a relation between the number and the type of sequons in glycoproteins.
Sequon types in viral envelope glycoproteins
The HA of influenza A H3N2 has accumulated many sequons (on average two additional sequons for NXS and three for NXT) in recent years between 1962 and 2009 (supplementary materialFigure S1 A and B, S2 A and B) [11]. By contrast, the number of sequons in gp120 of HIV-1 has remained relatively same over the years from 1981 to 2009 (Figure S1 C and D). It has been believed that the current number (˜26) of sequons in gp120 is the upper limit of sequons for this protein [11,20]. Interestingly, due to very high mutation rate in gp120 [10], there were considerable fluctuations in the sequon types with many disappearing and appearing time and again during the tracking period (Figure S2 C and D). As it may be seen (Figure S1 E to H) some sequon types are highly over-represented in these two envelope glycoproteins. This is because, given the low number of sequons, only a few sequon types are possible in these proteins (especially in HA) [13– 15]. However, as HIV-1 is known to have very high mutation rate [10], almost all sequon types are represented in gp120 during the period from 1981 to 2009. Our aim was to find whether the over-represented sequons in HA and gp120 are preferred sequon types or the result of random genetic drift [14]. As it is evident (see supplementary Table 1, Figure S1 E to H), the majority of the over-represented sequons in these two proteins are indeed preferred sequon types. This may provide some selective advantage for the viral envelope glycoproteins [13–,15],[20].
Conclusion
Eukaryotic N-glycoproteins do appear to show a preference for some types (based on the central amino acid X) of NXS/T sequons over others. The Markov chain analysis of NXS/T tri-peptide sequences in viral, archaeal and eukaryotic proteins also showed a similar preference indicating that this preference is not unique to N-glycoproteins. Not all possible NXS/T sequon types were generally seen in any single protein sequence. This may be partly due to the bias towards some sequon types, but also explained on the basis of probability sampling. The over-represented sequons found in viral envelope glycoproteins (HA of influenza A H3N2 and gp120 of HIV- 1) mostly represented the preferred NXS/T sequon types which might give them some selective advantage and hence are not entirely the result of random genetic drift.
Supplementary material
Figure 1.
Sequon types in Proteins. The mean number of sequon types versus the number of sequons per protein is shown for viral (A), archaeal (B) and eukaryotic (C) proteins, and experimentally confirmed eukaryotic N-glycoproteins (D). The NXS sequons are shown in red and NXT in blue. Inclined lines indicate the maximum number of sequon types and green dots show the mean number of sequon types per given number of sequons based on probability sampling
Acknowledgments
Authors thank Ole Thomsen Buus from Aarhus University for suggestions in computations.
Footnotes
Citation:Raoetal; Bioinformation 5(5): 208-212 (2010)
References
- 1.Spiro RG, et al. Glycobiol. 2002;12:43R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 2.Yan A, Lennarz WJ. J Biol Chem. 2005;280:3121. doi: 10.1074/jbc.R400036200. [DOI] [PubMed] [Google Scholar]
- 3.Gavel Y, von Heijne G. Protein Eng. 1990;3:433. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler R, et al. Biochim Biophys Acta. 1999;1473:4. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Dor S, et al. Glycobiol. 2004;14:95. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- 6.Petrescu AJ, et al. Glycobiol. 2004;14:103. [Google Scholar]
- 7.Kasturi L, et al. Biochem J. 1997;323:415. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy A, et al. Glycobiol. 1999;9:547. doi: 10.1093/glycob/9.6.547. [DOI] [PubMed] [Google Scholar]
- 9.Abu-Qarn M, Eichler J. Archaea. 2006;2:73-81. doi: 10.1155/2006/510578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belair M, et al. Aids Res Hum Retroviruses. 2009;25:199. doi: 10.1089/aid.2008.0162. [DOI] [PubMed] [Google Scholar]
- 11.Cui J, et al. Proc Natl Acad Sci USA. 2009;106:13421. doi: 10.1073/pnas.0905818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowarik M, et al. EMBO J. 2006;25:1957. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bush RM, et al. Mol Biol Evol. 1999;16:1457. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 14.Cherry JL, et al. Cherry. 2009;18:RRN1001. [Google Scholar]
- 15.Romanova J, et al. Virol. 2003;307:90. doi: 10.1016/s0042-6822(02)00064-8. [DOI] [PubMed] [Google Scholar]
- 16. http://www.uniprot.org/uniprot/?query=reviewed%3Ayes.
- 17. http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=database.
- 18. http://www.hiv.lanl.gov/components/sequence/HIV/search/search.html.
- 19.Rao RSP, et al. Bioinf Biol Insights. 2010;4:9. doi: 10.4137/bbi.s4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao RSP, Wollenweber B. Int J Biol Sci. 2010;6:407. doi: 10.7150/ijbs.6.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.