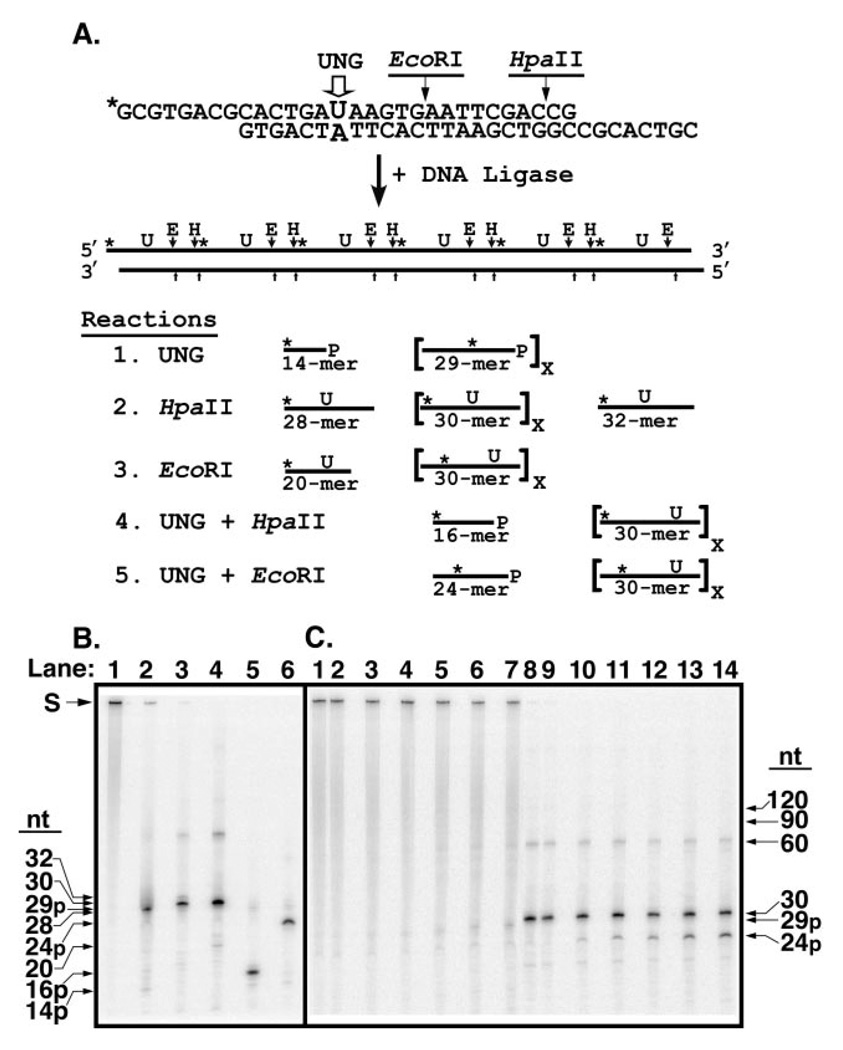
Fig. 7. Analysis of reaction products generated by UNG from a concatenated uracil-containing [32P]DNA substrate.
A, the concatenated uracil-containing [32P]DNA substrate was prepared by ligation of double-stranded 30-mer units as described under “Experimental Procedures.” Each 30-mer unit contained a uracil residue at position 15 in the upper strand opposite adenine in the lower, complementary, DNA strand; only the uracil-containing strand was 5′-32P-end labeled (marked with an asterisk, *). An EcoRI restriction endonuclease recognition site was situated 3′ to nucleotide 20 of the uracil-containing DNA strand. The 30-mer unit was designed with self-complementary overhanging 5′-ends of eight nucleotides that formed a HpaII endonuclease recognition site upon ligation. Ligation of the 30-mer units created a concatenated uracil-containing [32P]DNA substrate in which each uracil residue as well as the EcoRI and HpaII recognition sites were separated by 30 nucleotides. Reaction 1: sequential uracil excision by UNG followed by alkaline hydrolysis of the resultant AP-site produces 29-mers containing an internal 32P label and a 3′-phosphate (29p-mer). Excision of the 5′-terminal uracil produces a 5′-32P-end labeled 14-mer with a 3′-phosphate (14p-mer). Reaction 2: HpaII digestion of the concatenated [32P]DNA substrate and subsequent alkaline hydrolysis generates 30-mers with an internal 32P label. Cleavage by HpaII at the 5′- and 3′-ends yields a 28-mer containing a 5′-32P-end label and a 32-mer containing an internal 32P label, respectively. Reaction 3: EcoRI digestion produces 30-mers except at the 5′ terminus where a [32P]DNA-20-mer is generated. Reaction 4: concatenated [32P]DNA digested with UNG, then exhaustively digested with HpaII followed by alkaline hydrolysis, yields 16-mers containing a 3′-phosphate (16p-mer) where the uracil has been excised, but [32P]30-mers from intact uracil-containing 30-mer units, as shown in Reaction 2. Only 32P-labeled reaction products are shown. Reaction 5: concatenated [32P]DNA digested with UNG, then exhaustively digested with EcoRI followed by alkaline hydrolysis, yields [32P]24-mers containing a 3′-phosphate (24p-mer) where the uracil has been excised, but [32P]30-mers from uracil-containing 30-mer units, as shown in Reaction 3. Only 32P-labeled reaction products are shown. B, characterization of the concatenated uracil-containing [32P]DNA substrate. Reaction mixtures (10 µl) containing 10 pmol of concatenated [32P]DNA and no addition (lane 1), 70 units of UNG (lane 2), 10 units of EcoRI (lane 3), 10 units of HpaII (lane 4), 70 units of UNG and 10 units of HpaII (lane 5), or 70 units of UNG and 10 units of EcoRI (lane 6), were incubated at 37 °C, subjected to alkaline hydrolysis, and analyzed by 12% denaturing polyacrylamide gel electrophoresis as described under “Experimental Procedures.” The mobility of the unreacted concatenated [32P]DNA substrate (S) and various oligonucleotide reaction products (32-, 30-, 29p-, 28-, 24p-, 20-, 16p-, and 14p-mer) is indicated by arrows. C, time course of UNG digestion. A reaction mixture (100 µl) containing 0.06 units of UNG and 80 pmol of concatenated uracil-containing [32P]DNA was incubated at 37 °C. Aliquots (10 µl) were removed at 0, 5, 10, 20, 40, and 80 min and terminated by the addition of Ugi as described under “Experimental Procedures.” A control reaction contained 10 pmol of the [32P]DNA substrate only. Following reaction termination, each time point was divided into two aliquots. One, designated the UNG reaction, was subjected to alkaline hydrolysis. The other, designated as the EcoRI reaction, underwent EcoRI digestion and subsequent alkaline hydrolysis. Reactions were analyzed by denaturing 12% polyacrylamide, 8.3 m urea gel electrophoresis as described under “Experimental Procedures.” UNG reactions corresponding to the control reaction, and 0-, 5-, 10-, 20-, 40-, and 80-min time points were loaded in lanes 1–7, respectively, whereas the corresponding EcoRI reactions were loaded in lanes 8–14. Arrows indicate the positions of the various oligonucleotide reaction products. The extent of uracil excision after each incubation time was determined by dividing the amount of [32P]24p-mer oligonucleotide released by EcoRI digestion by the sum of [32P]24p-mer + [32P]30-mer, and multiplying the quotient by 100; this percentage was called the Extent of Digestion.