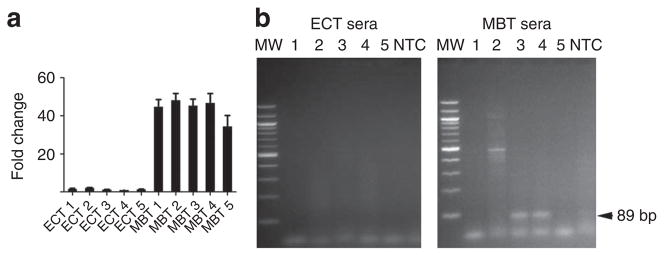
Figure 3. Detection of amplified c-Myc sequences in serum microvesicles from tumour-bearing mice.
M edulloblastoma (MBT; D425) and epidermoid carcinoma (ECT; A431) cells were used to generate subcutaneous tumours with and without c-Myc amplification, respectively. (a) c-Myc amplification was evaluated on all tumour samples at the RNA level after tumour resection. Values were normalized to GAPDH, presented as fold change compared with epidermoid carcinoma and shown as mean ± s.e.m. (n = 3). (b) ExoRNA was extracted from serum samples from five MBT and five ECT (MBT 1–5 and ECT 1–5, respectively). c-Myc PCR product was amplified using human specific primers. Amplified DNA was resolved by electrophoresis in a 2% agarose gel and visualized with ethidium bromide staining. c-Myc is shown as an 89 bp fragment (arrow). MW, molecular weight; NTC, no template control.